Developing high-affinity, oxygen-insensitive [NiFe]-hydrogenases as biocatalysts for energy conversion
- PMID: 37743798
- PMCID: PMC10657181
- DOI: 10.1042/BST20230120
Developing high-affinity, oxygen-insensitive [NiFe]-hydrogenases as biocatalysts for energy conversion
Abstract
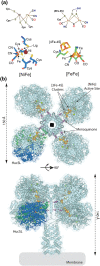
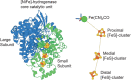
The splitting of hydrogen (H2) is an energy-yielding process, which is important for both biological systems and as a means of providing green energy. In biology, this reaction is mediated by enzymes called hydrogenases, which utilise complex nickel and iron cofactors to split H2 and transfer the resulting electrons to an electron-acceptor. These [NiFe]-hydrogenases have received considerable attention as catalysts in fuel cells, which utilise H2 to produce electrical current. [NiFe]-hydrogenases are a promising alternative to the platinum-based catalysts that currently predominate in fuel cells due to the abundance of nickel and iron, and the resistance of some family members to inhibition by gases, including carbon monoxide, which rapidly poison platinum-based catalysts. However, the majority of characterised [NiFe]-hydrogenases are inhibited by oxygen (O2), limiting their activity and stability. We recently reported the isolation and characterisation of the [NiFe]-hydrogenase Huc from Mycobacterium smegmatis, which is insensitive to inhibition by O2 and has an extremely high affinity, making it capable of oxidising H2 in air to below atmospheric concentrations. These properties make Huc a promising candidate for the development of enzyme-based fuel cells (EBFCs), which utilise H2 at low concentrations and in impure gas mixtures. In this review, we aim to provide context for the use of Huc for this purpose by discussing the advantages of [NiFe]-hydrogenases as catalysts and their deployment in fuel cells. We also address the challenges associated with using [NiFe]-hydrogenases for this purpose, and how these might be overcome to develop EBFCs that can be deployed at scale.
Keywords: [NiFe]-hydrogenase; electrocatalysis; enzyme-based fuel cells; hydrogen.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


References
-
- Nishikawa, K., Ogata, H. and Higuchi, Y. (2020) Structural basis of the function of [NiFe]-hydrogenases. Chem. Lett. 49, 164–173 10.1246/cl.190814 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

