In-depth Phylogenomic Analysis of Arbuscular Mycorrhizal Fungi Based on a Comprehensive Set of de novo Genome Assemblies
- PMID: 37744125
- PMCID: PMC10512289
- DOI: 10.3389/ffunb.2021.716385
In-depth Phylogenomic Analysis of Arbuscular Mycorrhizal Fungi Based on a Comprehensive Set of de novo Genome Assemblies
Abstract
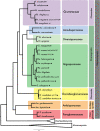
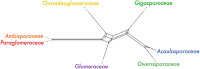
Morphological characters and nuclear ribosomal DNA (rDNA) phylogenies have so far been the basis of the current classifications of arbuscular mycorrhizal (AM) fungi. Improved understanding of the evolutionary history of AM fungi requires extensive ortholog sampling and analyses of genome and transcriptome data from a wide range of taxa. To circumvent the need for axenic culturing of AM fungi we gathered and combined genomic data from single nuclei to generate de novo genome assemblies covering seven families of AM fungi. We successfully sequenced the genomes of 15 AM fungal species for which genome data was not previously available. Comparative analysis of the previously published Rhizophagus irregularis DAOM197198 assembly confirm that our novel workflow generates genome assemblies suitable for phylogenomic analysis. Predicted genes of our assemblies, together with published protein sequences of AM fungi and their sister clades, were used for phylogenomic analyses. We evaluated the phylogenetic placement of Glomeromycota in relation to its sister phyla (Mucoromycota and Mortierellomycota), and found no support to reject a polytomy. Finally, we explored the phylogenetic relationships within Glomeromycota. Our results support family level classification from previous phylogenetic studies, and the polyphyly of the order Glomerales with Claroideoglomeraceae as the sister group to Glomeraceae and Diversisporales.
Keywords: Glomeromycota; genomics; phylogenetic; single nuclei sequencing; topology.
Copyright © 2021 Montoliu-Nerin, Sánchez-García, Bergin, Kutschera, Johannesson, Bever and Rosling.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Beaudet D., Chen E. C., Mathieu S., Yildirir G., Ndikumana S., Dalpé Y., et al. . (2018). Ultra-low input transcriptomics reveal the spore functional content and phylogenetic affiliations of poorly studied arbuscular mycorrhizal fungi. DNA Res. 25, 217–227. 10.1093/dnares/dsx051 - DOI - PMC - PubMed
-
- Bonfante P., Venice F. (2020). Mucoromycota: going to the roots of plant-interacting fungi. Fungal Biol. Rev. 34, 100–113. 10.1016/j.fbr.2019.12.003 - DOI
LinkOut - more resources
Full Text Sources

