Natural and anthropogenic carbon input affect microbial activity in salt marsh sediment
- PMID: 37744927
- PMCID: PMC10512730
- DOI: 10.3389/fmicb.2023.1235906
Natural and anthropogenic carbon input affect microbial activity in salt marsh sediment
Abstract
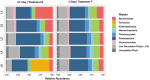
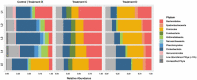
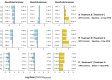
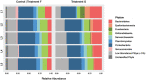
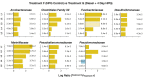
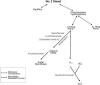
Salt marshes are dynamic, highly productive ecosystems positioned at the interface between terrestrial and marine systems. They are exposed to large quantities of both natural and anthropogenic carbon input, and their diverse sediment-hosted microbial communities play key roles in carbon cycling and remineralization. To better understand the effects of natural and anthropogenic carbon on sediment microbial ecology, several sediment cores were collected from Little Sippewissett Salt Marsh (LSSM) on Cape Cod, MA, USA and incubated with either Spartina alterniflora cordgrass or diesel fuel. Resulting shifts in microbial diversity and activity were assessed via bioorthogonal non-canonical amino acid tagging (BONCAT) combined with fluorescence-activated cell sorting (FACS) and 16S rRNA gene amplicon sequencing. Both Spartina and diesel amendments resulted in initial decreases of microbial diversity as well as clear, community-wide shifts in metabolic activity. Multi-stage degradative frameworks shaped by fermentation were inferred based on anabolically active lineages. In particular, the metabolically versatile Marinifilaceae were prominent under both treatments, as were the sulfate-reducing Desulfovibrionaceae, which may be attributable to their ability to utilize diverse forms of carbon under nutrient limited conditions. By identifying lineages most directly involved in the early stages of carbon processing, we offer potential targets for indicator species to assess ecosystem health and highlight key players for selective promotion of bioremediation or carbon sequestration pathways.
Keywords: biogeochemistry; carbon cycle; metabolic activity; microbial diversity; salt marsh.
Copyright © 2023 Frates, Spietz, Silverstein, Girguis, Hatzenpichler and Marlow.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures






Similar articles
-
Estuarine Sediment Microbiomes from a Chronosequence of Restored Urban Salt Marshes.Microb Ecol. 2023 Apr;85(3):916-930. doi: 10.1007/s00248-023-02193-y. Epub 2023 Feb 24. Microb Ecol. 2023. PMID: 36826588
-
The core root microbiome of Spartina alterniflora is predominated by sulfur-oxidizing and sulfate-reducing bacteria in Georgia salt marshes, USA.Microbiome. 2022 Mar 1;10(1):37. doi: 10.1186/s40168-021-01187-7. Microbiome. 2022. PMID: 35227326 Free PMC article.
-
Salt Marsh Bacterial Communities before and after the Deepwater Horizon Oil Spill.Appl Environ Microbiol. 2017 Sep 29;83(20):e00784-17. doi: 10.1128/AEM.00784-17. Print 2017 Oct 15. Appl Environ Microbiol. 2017. PMID: 28778895 Free PMC article.
-
Soil microbial community development across a 32-year coastal wetland restoration time series and the relative importance of environmental factors.Sci Total Environ. 2022 May 15;821:153359. doi: 10.1016/j.scitotenv.2022.153359. Epub 2022 Jan 23. Sci Total Environ. 2022. PMID: 35081409
-
The Minderoo-Monaco Commission on Plastics and Human Health.Ann Glob Health. 2023 Mar 21;89(1):23. doi: 10.5334/aogh.4056. eCollection 2023. Ann Glob Health. 2023. PMID: 36969097 Free PMC article. Review.
References
-
- Ahmed S., Krumpelt M., Pereira C., Wilkenhoener R. (1999). Liquid fuel reformer development (No. ANL/CMT/CP-99684). Lemont, IL: Argonne National Lab.
-
- Aitken M. D., Stringfellow W. T., Nagel R. D., Kazunga C., Chen S. H. (1998). Characteristics of phenanthrene-degrading bacteria isolated from soils contaminated with polycyclic aromatic hydrocarbons. Can. J. Microbiol. 44 743–752. - PubMed
-
- Bagert J. D., Xie Y. J., Sweredoski M. J., Qi Y., Hess S., Schuman E. M., et al. (2014). Quantitative, time-resolved proteomic analysis by combining bioorthogonal noncanonical amino acid tagging and pulsed stable isotope labeling by amino acids in cell culture. Mol. Cell. Proteom. 13 1352–1358. 10.1074/mcp.M113.031914 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

