Molecular characterization and differential effects of levofloxacin and ciprofloxacin on the potential for developing quinolone resistance among clinical Pseudomonas aeruginosa isolates
- PMID: 37744929
- PMCID: PMC10514475
- DOI: 10.3389/fmicb.2023.1209224
Molecular characterization and differential effects of levofloxacin and ciprofloxacin on the potential for developing quinolone resistance among clinical Pseudomonas aeruginosa isolates
Abstract
Background: Fluoroquinolones are some of the most used antimicrobial agents for the treatment of Pseudomonas aeruginosa. This study aimed at exploring the differential activity of ciprofloxacin and levofloxacin on the selection of resistance among P. aeruginosa isolates at our medical center.
Methods: 233 P. aeruginosa clinical isolates were included in this study. Antimicrobial susceptibility testing (AST) was done using disk diffusion and broth microdilution assays. Random Amplification of Polymorphic DNA (RAPD) was done to determine the genetic relatedness between the isolates. Induction of resistance against ciprofloxacin and levofloxacin was done on 19 isolates. Fitness cost assay was done on the 38 induced mutants and their parental isolates. Finally, whole genome sequencing was done on 16 induced mutants and their 8 parental isolates.
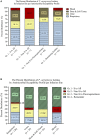
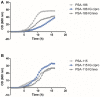
Results: AST results showed that aztreonam had the highest non-susceptibility. RAPD results identified 18 clusters. The 19 P. aeruginosa isolates that were induced against ciprofloxacin and levofloxacin yielded MICs ranging between 16 and 256 μg/mL. Levofloxacin required fewer passages in 10 isolates and the same number of passages in 9 isolates as compared to ciprofloxacin to reach their breakpoints. Fitness cost results showed that 12 and 10 induced mutants against ciprofloxacin and levofloxacin, respectively, had higher fitness cost when compared to their parental isolates. Whole genome sequencing results showed that resistance to ciprofloxacin and levofloxacin in sequenced mutants were mainly associated with alterations in gyrA, gyrB and parC genes.
Conclusion: Understanding resistance patterns and risk factors associated with infections is crucial to decrease the emerging threat of antimicrobial resistance.
Keywords: Pseudomonas aeruginosa; antimicrobial resistance; fitness cost; fluoroquinolone resistance; hospital-acquired infection; nosocomial infection.
Copyright © 2023 Kanafani, Sleiman, Frem, Doumat, Gharamti, El Hafi, Doumith, AlGhoribi, Kanj, Araj, Matar and Abou Fayad.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
[Susceptibility of 570 Pseudomonas aeruginosa strains to 11 antimicrobial agents and the mechanism of its resistance to fluoroquinolones].Zhonghua Yi Xue Za Zhi. 2003 Mar 10;83(5):403-7. Zhonghua Yi Xue Za Zhi. 2003. PMID: 12820918 Chinese.
-
The role of gyrA and parC mutations in fluoroquinolones-resistant Pseudomonas aeruginosa isolates from Iran.Braz J Microbiol. 2016 Oct-Dec;47(4):925-930. doi: 10.1016/j.bjm.2016.07.016. Epub 2016 Jul 26. Braz J Microbiol. 2016. PMID: 27522930 Free PMC article.
-
Fluoroquinolone resistance of Pseudomonas aeruginosa isolates causing nosocomial infection is correlated with levofloxacin but not ciprofloxacin use.Int J Antimicrob Agents. 2010 Mar;35(3):261-4. doi: 10.1016/j.ijantimicag.2009.11.007. Epub 2009 Dec 31. Int J Antimicrob Agents. 2010. PMID: 20045290
-
Fluoroquinolone resistance contributing mechanisms and genotypes of ciprofloxacin- unsusceptible Pseudomonas aeruginosa strains in Iran: emergence of isolates carrying qnr/aac(6)-Ib genes.Int Microbiol. 2022 Aug;25(3):405-415. doi: 10.1007/s10123-021-00220-x. Epub 2021 Oct 28. Int Microbiol. 2022. PMID: 34709520
-
Ciprofloxacin resistance and tolerance of Pseudomonas aeruginosa ocular isolates.Cont Lens Anterior Eye. 2023 Jun;46(3):101819. doi: 10.1016/j.clae.2023.101819. Epub 2023 Feb 1. Cont Lens Anterior Eye. 2023. PMID: 36732125
Cited by
-
Uncovering Microbial Composition of the Tissue Microenvironment in Bladder Cancer using RNA Sequencing Data.J Cancer. 2024 Mar 4;15(8):2431-2441. doi: 10.7150/jca.93055. eCollection 2024. J Cancer. 2024. PMID: 38495492 Free PMC article.
-
Selective Serotonin Reuptake Inhibitors: Antimicrobial Activity Against ESKAPEE Bacteria and Mechanisms of Action.Antibiotics (Basel). 2025 Jan 8;14(1):51. doi: 10.3390/antibiotics14010051. Antibiotics (Basel). 2025. PMID: 39858337 Free PMC article.
-
Molecular Characteristics of Colistin Resistance in Acinetobacter baumannii and the Activity of Antimicrobial Combination Therapy in a Tertiary Care Medical Center in Lebanon.Microorganisms. 2024 Feb 8;12(2):349. doi: 10.3390/microorganisms12020349. Microorganisms. 2024. PMID: 38399753 Free PMC article.
-
Comparing antibiotic resistance and virulence profiles of Enterococcus faecium, Klebsiella pneumoniae, and Pseudomonas aeruginosa from environmental and clinical settings.Heliyon. 2024 Apr 26;10(9):e30215. doi: 10.1016/j.heliyon.2024.e30215. eCollection 2024 May 15. Heliyon. 2024. PMID: 38720709 Free PMC article.
-
Multidrug-Resistant Commensal and Infection-Causing Staphylococcus spp. Isolated from Companion Animals in the Valencia Region.Vet Sci. 2024 Jan 26;11(2):54. doi: 10.3390/vetsci11020054. Vet Sci. 2024. PMID: 38393072 Free PMC article.
References
LinkOut - more resources
Full Text Sources

