An improved framework for detecting discrete epidemiologically meaningful partitions in hierarchically clustered genetic data
- PMID: 37744999
- PMCID: PMC10517639
- DOI: 10.1093/bioadv/vbad118
An improved framework for detecting discrete epidemiologically meaningful partitions in hierarchically clustered genetic data
Abstract
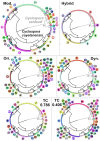
Motivation: Hierarchical clustering of microbial genotypes has the limitation that hierarchical clusters are nested, where smaller groups of related isolates exist within larger groups that get progressively larger as relationships become increasingly distant. In an epidemiologic context, investigators must dissect hierarchical trees into discrete groupings that are epidemiologically meaningful. We recently described a statistical framework (Method A) for dissecting hierarchical trees that attempts to minimize investigator bias. Here, we apply a modified version of that framework (Method B) to a hierarchical tree constructed from 2111 genotypes of the foodborne parasite Cyclospora, including 639 genotypes linked to epidemiologically defined outbreaks. To evaluate Method B's performance, we examined the concordance between these epidemiologically defined groupings and the genetic partitions identified. We also used the same epidemiologic clusters to evaluate the performance of Method A, plus two tree-dissection methods (cutreeHybrid and cutreeDynamic) available within the Dynamic Tree Cut R package, in addition to the TreeCluster method and PARNAS.
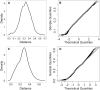
Results: Compared to the other methods, Method B, TreeCluster, and PARNAS were the most accurate (99.4%) in identifying genetic groups that reflected the epidemiologic groupings, noting that TreeCluster and PARNAS performed identically on our dataset. CutreeHybrid identified groups reflecting patterns in the wider Cyclospora population structure but lacked finer, strain-level discrimination (Simpson's D: cutreeHybrid=0.785). CutreeDynamic displayed good strain discrimination (Simpson's D = 0.933), though lacked sensitivity (77%). At two different threshold/radius settings TreeCluster/PARNAS displayed similar utility to Method B. However, Method B computes a tree-dissection threshold automatically, and the threshold/radius settings used when executing TreeCluster/PARNAS here were computed using Method B. Using a TreeCluster threshold of 0.045 as recommended in the TreeCluster documentation, epidemiologic utility dropped markedly below that of Method B.
Availability and implementation: Relevant code and data are publicly available. Source code (Method B) and instructions for its use are available here: https://github.com/Joel-Barratt/Hierarchical-tree-dissection-framework.
Published by Oxford University Press 2023.
Conflict of interest statement
None declared.
Figures

References
-
- Anonymous. Domestically acquired cases of cyclosporiasis—United States, May–August 2018. Centers for Disease Control and Prevention, 2018.
-
- Anonymous. Domestically acquired cases of cyclosporiasis—United States, May–August 2019. Centers for Disease Control and Prevention, 2019a.
-
- Anonymous. Outbreak of cyclospora infections linked to fresh basil from Siga Logistics de RL de CV of Morelos, Mexico. Centers for Disease Control and Prevention, 2019b.
-
- Anonymous. Domestically acquired cases of cyclosporiasis—United States, May–August 2020. Centers for Disease Control and Prevention, 2020.
LinkOut - more resources
Full Text Sources

