ChromNetMotif: a Python tool to extract chromatin-sate marked motifs in a chromatin interaction network
- PMID: 37745003
- PMCID: PMC10517636
- DOI: 10.1093/bioadv/vbad126
ChromNetMotif: a Python tool to extract chromatin-sate marked motifs in a chromatin interaction network
Abstract
Motivation: Analysis of network motifs is crucial to studying the robustness, stability, and functions of complex networks. Genome organization can be viewed as a biological network that consists of interactions between different chromatin regions. These interacting regions are also marked by epigenetic or chromatin states which can contribute to the overall organization of the chromatin and proper genome function. Therefore, it is crucial to integrate the chromatin states of the nodes when performing motif analysis in chromatin interaction networks. Even though there has been increasing production of chromatin interaction and genome-wide epigenetic modification data, there is a lack of publicly available tools to extract chromatin state-marked motifs from genome organization data.
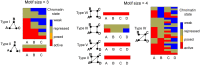
Results: We develop a Python tool, ChromNetMotif, offering an easy-to-use command line interface to extract chromatin-state-marked motifs from a chromatin interaction network. The tool can extract occurrences, frequencies, and statistical enrichment of the chromatin state-marked motifs. Visualization files are also generated which allow the user to interpret the motifs easily. ChromNetMotif also allows the user to leverage the features of a multicore processor environment to reduce computation time for larger networks. The output files generated can be used to perform further downstream analysis. ChromNetMotif aims to serve as an important tool to comprehend the interplay between epigenetics and genome organization.
Availability and implementation: ChromNetMotif is available at https://github.com/lncRNAAddict/ChromNetworkMotif.
© The Author(s) 2023. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures

References
-
- Angulo MT, Liu Y-Y, Slotine J-J. et al. Network motifs emerge from interconnections that favour stability. Nature Phys 2015;11:848–52.
Grants and funding
LinkOut - more resources
Full Text Sources

