Single-cell and genetic multi-omics analysis combined with experiments confirmed the signature and potential targets of cuproptosis in hepatocellular carcinoma
- PMID: 37745297
- PMCID: PMC10516581
- DOI: 10.3389/fcell.2023.1240390
Single-cell and genetic multi-omics analysis combined with experiments confirmed the signature and potential targets of cuproptosis in hepatocellular carcinoma
Abstract
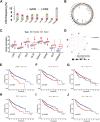
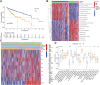
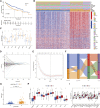
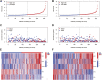
Background: Cuproptosis, as a recently discovered type of programmed cell death, occupies a very important role in hepatocellular carcinoma (HCC) and provides new methods for immunotherapy; however, the functions of cuproptosis in HCC are still unclear. Methods: We first analyzed the transcriptome data and clinical information of 526 HCC patients using multiple algorithms in R language and extensively described the copy number variation, prognostic and immune infiltration characteristics of cuproptosis related genes (CRGs). Then, the hub CRG related genes associated with prognosis through LASSO and Cox regression analyses and constructed a prognostic prediction model including multiple molecular markers and clinicopathological parameters through training cohorts, then this model was verified by test cohorts. On the basis of the model, the clinicopathological indicators, immune infiltration and tumor microenvironment characteristics of HCC patients were further explored via bioinformation analysis. Then, We further explored the key gene biological function by single-cell analysis, cell viability and transwell experiments. Meantime, we also explored the molecular docking of the hub genes. Results: We have screened 5 hub genes associated with HCC prognosis and constructed a prognosis prediction scoring model. And the model results showed that patients in the high-risk group had poor prognosis and the expression levels of multiple immune markers, including PD-L1, CD276 and CTLA4, were higher than those patients in the low-risk group. We found a significant correlation between risk score and M0 macrophages and memory CD4+ T cells. And the single-cell analysis and molecular experiments showed that BEX1 were higher expressed in HCC tissues and deletion inhibited the proliferation, invasion and migration and EMT pathway of HCC cells. Finally, it was observed that BEX1 could bind to sorafenib to form a stable conformation. Conclusion: The study not only revealed the multiomics characteristics of CRGs in HCC but also constructed a new high-accuracy prognostic prediction model. Meanwhile, BEX1 were also identified as hub genes that can mediate the cuproptosis of hepatocytes as potential therapeutic targets for HCC.
Keywords: cuproptosis; hepatocellular carcinoma; immune microenvironment; molecular docking; programmed cell death; single-cell RNA-sequencing.
Copyright © 2023 Cao, Qi, Wu, Li and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Identification of cuproptosis-related subtypes, construction of a prognosis model, and tumor microenvironment landscape in gastric cancer.Front Immunol. 2022 Nov 21;13:1056932. doi: 10.3389/fimmu.2022.1056932. eCollection 2022. Front Immunol. 2022. PMID: 36479114 Free PMC article.
-
Comprehensive analysis of cuproptosis-related prognostic gene signature and tumor immune microenvironment in HCC.Front Genet. 2023 Feb 20;14:1094793. doi: 10.3389/fgene.2023.1094793. eCollection 2023. Front Genet. 2023. PMID: 36891150 Free PMC article.
-
Cuproptosis-Related Signature Predicts the Prognosis, Tumor Microenvironment, and Drug Sensitivity of Hepatocellular Carcinoma.J Immunol Res. 2022 Nov 16;2022:3393027. doi: 10.1155/2022/3393027. eCollection 2022. J Immunol Res. 2022. PMID: 36438201 Free PMC article.
-
Identification of a novel cuproptosis-related gene signature and integrative analyses in patients with lower-grade gliomas.Front Immunol. 2022 Aug 15;13:933973. doi: 10.3389/fimmu.2022.933973. eCollection 2022. Front Immunol. 2022. PMID: 36045691 Free PMC article. Review.
-
Cuproptosis and cuproptosis-related cell death and genes: mechanistic links to spermatogenic cell death.Cell Death Discov. 2025 Jun 10;11(1):274. doi: 10.1038/s41420-025-02553-2. Cell Death Discov. 2025. PMID: 40494847 Free PMC article. Review.
Cited by
-
Identification of ASF1A and HJURP by global H3-H4 histone chaperone analysis as a prognostic two-gene model in hepatocellular carcinoma.Sci Rep. 2024 Apr 1;14(1):7666. doi: 10.1038/s41598-024-58368-1. Sci Rep. 2024. PMID: 38561384 Free PMC article.
References
-
- Cai J., Chen L., Zhang Z., Zhang X., Lu X., Liu W., et al. (2019). Genome-wide mapping of 5-hydroxymethylcytosines in circulating cell-free DNA as a non-invasive approach for early detection of hepatocellular carcinoma. Multicent. Study 68 (12), 2195–2205. 10.1136/gutjnl-2019-318882 - DOI - PMC - PubMed
-
- Cheng A. L., Kang Y. K., Chen Z., Tsao C. J., Qin S., Kim J. S., et al. (2009). Efficacy and safety of sorafenib in patients in the asia-pacific region with advanced hepatocellular carcinoma: A phase III randomised, double-blind, placebo-controlled trial. LANCET Oncol. 10 (1), 25–34. 10.1016/S1470-2045(08)70285-7 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

