Bioinformatics-based analysis reveals elevated CYTL1 as a potential therapeutic target for BRAF-mutated melanoma
- PMID: 37745303
- PMCID: PMC10516578
- DOI: 10.3389/fcell.2023.1171047
Bioinformatics-based analysis reveals elevated CYTL1 as a potential therapeutic target for BRAF-mutated melanoma
Abstract
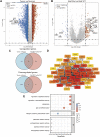
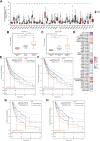
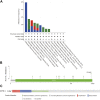
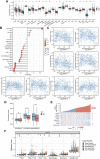
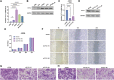
Introduction: Despite many recent emerging therapeutic modalities that have prolonged the survival of melanoma patients, the prognosis of melanoma remains discouraging, and further understanding of the mechanisms underlying melanoma progression is needed. Melanoma patients often have multiple genetic mutations, with BRAF mutations being the most common. In this study, public databases were exploited to explore a potential therapeutic target for BRAF-mutated melanoma. Methods: In this study, we analyzed differentially expressed genes (DEGs) in normal tissues and melanomas, Braf wild-type and Braf mutant melanomas using information from TCGA databases and the GEO database. Subsequently, we analyzed the differential expression of CYTL1 in various tumor tissues and its effect on melanoma prognosis, and resolved the mutation status of CYTL1 and its related signalling pathways. By knocking down CYTL1 in melanoma cells, the effects of CYTL1 on melanoma cell proliferation, migration and invasion were further examined by CCK8 assay, Transwell assay and cell migration assay. Results: 24 overlapping genes were identified by analyzing DEGs common to melanoma and normal tissue, BRAF-mutated and BRAF wild-type melanoma. Among them, CYTL1 was highly expressed in melanoma, especially in BRAF-mutated melanoma, and the high expression of CYTL1 was associated with epithelial-mesenchymal transition (EMT), cell cycle, and cellular response to UV. In melanoma patients, especially BRAF-mutated melanoma patients, clinical studies showed a positive correlation between increased CYTL1 expression and shorter overall survival (OS) and disease-free survival (DFS). In vitro experiments further confirmed that the knockdown of CYTL1 significantly inhibited the migration and invasive ability of melanoma cells. Conclusion: CYTL1 is a valuable prognostic biomarker and a potentially effective therapeutic target in melanoma, especially BRAF-mutated melanoma.
Keywords: BRAF mutations; CYTL1; cell migration and invasion; melanoma; molecular biomarker.
Copyright © 2023 Tao, Cui, Sun, Cao, Dai, Ge, Zhang, Ma and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2 (5), 401–404. 10.1158/2159-8290.CD-12-0095 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

