Identification and verification of a prognostic signature based on a miRNA-mRNA interaction pattern in colon adenocarcinoma
- PMID: 37745305
- PMCID: PMC10511881
- DOI: 10.3389/fcell.2023.1161667
Identification and verification of a prognostic signature based on a miRNA-mRNA interaction pattern in colon adenocarcinoma
Abstract
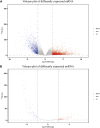
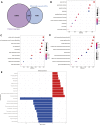
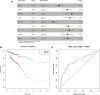
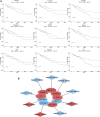
The expression characteristics of non-coding RNA (ncRNA) in colon adenocarcinoma (COAD) are involved in regulating various biological processes. To achieve these functions, ncRNA and a member of the Argonaute protein family form an RNA-induced silencing complex (RISC). The RISC is directed by ncRNA, especially microRNA (miRNA), to bind the target complementary mRNAs and regulate their expression by interfering with mRNA cleavage, degradation, or translation. However, how to identify potential miRNA biomarkers and therapeutic targets remains unclear. Here, we performed differential gene screening based on The Cancer Genome Atlas dataset and annotated meaningful differential genes to enrich related biological processes and regulatory cancer pathways. According to the overlap between the screened differential mRNAs and differential miRNAs, a prognosis model based on a least absolute shrinkage and selection operator-based Cox proportional hazards regression analysis can be established to obtain better prognosis characteristics. To further explore the therapeutic potential of miRNA as a target of mRNA intervention, we conducted an immunohistochemical analysis and evaluated the expression level in the tissue microarray of 100 colorectal cancer patients. The results demonstrated that the expression level of POU4F1, DNASE1L2, and WDR72 in the signature was significantly upregulated in COAD and correlated with poor prognosis. Establishing a prognostic signature based on miRNA target genes will help elucidate the molecular pathogenesis of COAD and provide novel potential targets for RNA therapy.
Keywords: RNA therapy; colon adenocarcinoma; miRNAs; prognostic signature; tissue microarray.
Copyright © 2023 Zhao, Li, Li, Guo, Jia, Xu, Chen, Shen and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources

