This is a preprint.
Competition and Synergy of Arp2/3 and Formins in Nucleating Actin Waves
- PMID: 37745345
- PMCID: PMC10515902
- DOI: 10.1101/2023.09.13.557508
Competition and Synergy of Arp2/3 and Formins in Nucleating Actin Waves
Update in
-
Competition and synergy of Arp2/3 and formins in nucleating actin waves.Cell Rep. 2024 Jul 23;43(7):114423. doi: 10.1016/j.celrep.2024.114423. Epub 2024 Jul 4. Cell Rep. 2024. PMID: 38968072 Free PMC article.
Abstract
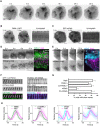
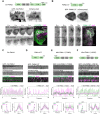
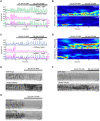
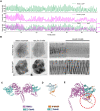
The assembly and disassembly of actin filaments and their regulatory proteins are crucial for maintaining cell structure or changing physiological state. However, because of the tremendous global impact of actin on diverse cellular processes, dissecting the specific role of actin regulatory proteins remains challenging. In this study, we employ actin waves that propagate on the cortex of mast cell to investigate the interplay between formins and the Arp2/3 complex in the nucleating and turnover of cortical actin. Our findings reveal that the recruitment of FMNL1 and mDia3 precedes the Arp2/3 complex in cortical actin waves. Membrane and GTPase-interaction can drive oscillations of FMNL1 in an actin-dependent manner, but active Cdc42 waves or constitutively-active FMNL1 mutant can form without actin waves. In addition to the apparent coordinated assembly of formins and Arp2/3, we further reveal their antagonism, where inhibition of Arp2/3 complex by CK-666 led to a transient increase in the recruitment of formins and actin polymerization. Our analysis suggest that the antagonism could not be explained for the competition between FMNL1 and Arp2/3 for monomeric actin. Rather, it is regulated by a limited pool of their common upstream regulator, Cdc42, whose level is negatively regulated by Arp2/3. Collectively, our study highlights the multifaceted interactions, cooperative or competitive, between formins and Arp2/3 complex, in the intricate and dynamic control of actin cytoskeletal network.
Figures

References
-
- Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Siminovitch KA, Rosen MK. 1999. Structure of Cdc42 in complex with the GTPase-binding domain of the “Wiskott–Aldrich syndrome” protein. Nature 399:379–383. - PubMed
-
- Bershadsky A. 2004. Magic touch: how does cell-cell adhesion trigger actin assembly? Trends Cell Biol 14:1–5. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
