This is a preprint.
Mapping the Single-cell Differentiation Landscape of Osteosarcoma
- PMID: 37745374
- PMCID: PMC10515803
- DOI: 10.1101/2023.09.13.555156
Mapping the Single-cell Differentiation Landscape of Osteosarcoma
Update in
-
Mapping the Single-Cell Differentiation Landscape of Osteosarcoma.Clin Cancer Res. 2024 Aug 1;30(15):3259-3272. doi: 10.1158/1078-0432.CCR-24-0563. Clin Cancer Res. 2024. PMID: 38775859 Free PMC article.
Abstract
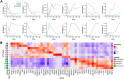
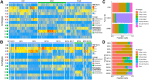
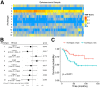
The genetic and intratumoral heterogeneity observed in human osteosarcomas (OS) poses challenges for drug development and the study of cell fate, plasticity, and differentiation, processes linked to tumor grade, cell metastasis, and survival. To pinpoint errors in OS differentiation, we transcriptionally profiled 31,527 cells from a tissue-engineered model that directs MSCs toward adipogenic and osteoblastic fates. Incorporating pre-existing chondrocyte data, we applied trajectory analysis and non-negative matrix factorization (NMF) to generate the first human mesenchymal differentiation atlas. This 'roadmap' served as a reference to delineate the cellular composition of morphologically complex OS tumors and quantify each cell's lineage commitment. Projecting these signatures onto a bulk RNA-seq OS dataset unveiled a correlation between a stem-like transcriptomic phenotype and poorer survival outcomes. Our study takes the critical first step in accurately quantifying OS differentiation and lineage, a prerequisite to better understanding global differentiation bottlenecks that might someday be targeted therapeutically.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Jo VY, Doyle LA. Refinements in Sarcoma Classification in the Current 2013 World Health Organization Classification of Tumours of Soft Tissue and Bone. Surg Oncol Clin N Am 2016;25:621–43. - PubMed
-
- Thway K. Pathology of soft tissue sarcomas. Clin Oncol (R Coll Radiol) 2009;21:695–705. - PubMed
-
- Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer 2014;14:722–35. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
