This is a preprint.
Integration of Patient-Derived Organoids and Organ-on-Chip Systems: Investigating Colorectal Cancer Invasion within the Mechanical and GABAergic Tumor Microenvironment
- PMID: 37745376
- PMCID: PMC10515884
- DOI: 10.1101/2023.09.14.557797
Integration of Patient-Derived Organoids and Organ-on-Chip Systems: Investigating Colorectal Cancer Invasion within the Mechanical and GABAergic Tumor Microenvironment
Abstract
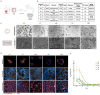
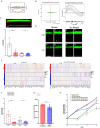
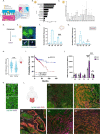
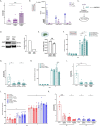
Three-dimensional (3D) in vitro models are essential in cancer research, but they often neglect physical forces. In our study, we combined patient-derived tumor organoids with a microfluidic organ-on-chip system to investigate colorectal cancer (CRC) invasion in the tumor microenvironment (TME). This allowed us to create patient-specific tumor models and assess the impact of physical forces on cancer biology. Our findings showed that the organoid-on-chip models more closely resembled patient tumors at the transcriptional level, surpassing organoids alone. Using 'omics' methods and live-cell imaging, we observed heightened responsiveness of KRAS mutant tumors to TME mechanical forces. These tumors also utilized the γ-aminobutyric acid (GABA) neurotransmitter as an energy source, increasing their invasiveness. This bioengineered model holds promise for advancing our understanding of cancer progression and improving CRC treatments.
Keywords: invasion; neurotransmitters; organoids; organs-on-chips; peristalsis.
Conflict of interest statement
DECLARATION OF INTEREST The authors declare no competing interest.
Figures

References
-
- Wakefield L., Agarwal S. & Tanner K. Preclinical models for drug discovery for metastatic disease. Cell 186, 1792–1813 (2023). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
