This is a preprint.
Transcriptional Signatures of Hippocampal Tau Pathology in Primary Age-Related Tauopathy and Alzheimer's Disease
- PMID: 37745408
- PMCID: PMC10516095
- DOI: 10.1101/2023.09.12.23295440
Transcriptional Signatures of Hippocampal Tau Pathology in Primary Age-Related Tauopathy and Alzheimer's Disease
Update in
-
Transcriptional signatures of hippocampal tau pathology in primary age-related tauopathy and Alzheimer's disease.Cell Rep. 2025 Mar 25;44(3):115422. doi: 10.1016/j.celrep.2025.115422. Epub 2025 Mar 13. Cell Rep. 2025. PMID: 40085647 Free PMC article.
Abstract
Background: Tau pathology is common in age-related neurodegenerative diseases. Tau pathology in primary age-related tauopathy (PART) and in Alzheimer's disease (AD) has a similar biochemical structure and anatomic distribution, which is distinct from tau pathology in other diseases. However, the molecular changes associated with intraneuronal tau pathology in PART and AD, and whether these changes are similar in the two diseases, is largely unexplored.
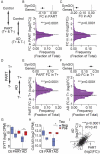
Methods: Using GeoMx spatial transcriptomics, mRNA was quantified in CA1 pyramidal neurons with tau pathology and adjacent neurons without tau pathology in 6 cases of PART and 6 cases of AD, and compared to 4 control cases without pathology. Transcriptional changes were analyzed for differential gene expression and for coordinated patterns of gene expression associated with both disease state and intraneuronal tau pathology.
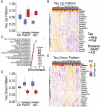
Results: Synaptic gene changes and two novel gene expression signatures associated with intraneuronal tau were identified in PART and AD. Overall, gene expression changes associated with intraneuronal tau pathology were similar in PART and AD. Synaptic gene expression was decreased overall in neurons in AD and PART compared to control cases. However, this decrease was largely driven by neurons lacking tau pathology. Synaptic gene expression was increased in tau-positive neurons compared to tau-negative neurons in disease. Two novel gene expression signatures associated with intraneuronal tau were identified by examining coordinated patterns of gene expression. Genes in the up-regulated expression pattern were enriched in calcium regulation and synaptic function pathways, specifically in synaptic exocytosis. These synaptic gene changes and intraneuronal tau expression signatures were confirmed in a published transcriptional dataset of cortical neurons with tau pathology in AD.
Conclusions: PART and AD show similar transcriptional changes associated with intraneuronal tau pathology in CA1 pyramidal neurons, raising the possibility of a mechanistic relationship between the tau pathology in the two diseases. Intraneuronal tau pathology was also associated with increased expression of genes associated with synaptic function and calcium regulation compared to tau-negative disease neurons. The findings highlight the power of molecular analysis stratified by pathology in neurodegenerative disease and provide novel insight into common molecular pathways associated with intraneuronal tau in PART and AD.
Conflict of interest statement
Competing Interests: The authors have no competing interests to declare.
Figures


References
-
- Pletnikova O, Kageyama Y, Rudow G, LaClair KD, Albert M, Crain BJ, Tian J, Fowler D, Troncoso JC: The spectrum of preclinical Alzheimer’s disease pathology and its modulation by ApoE genotype. Neurobiol Aging 2018, 71:72–80. - PubMed
-
- Braak H, Thal DR, Ghebremedhin E, Del Tredici K: Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J Neuropathol Exp Neurol 2011, 70:960–969. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
