This is a preprint.
Determinants of de novo B cell responses to drifted epitopes in post-vaccination SARS-CoV-2 infections
- PMID: 37745498
- PMCID: PMC10516057
- DOI: 10.1101/2023.09.12.23295384
Determinants of de novo B cell responses to drifted epitopes in post-vaccination SARS-CoV-2 infections
Update in
-
Intrinsic immunogenicity is a major determinant of type-specific responses in SARS-CoV-2 infections.Nat Immunol. 2025 Jun;26(6):829-836. doi: 10.1038/s41590-025-02162-2. Epub 2025 May 27. Nat Immunol. 2025. PMID: 40425779 Free PMC article.
Abstract
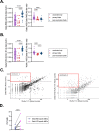
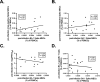
Vaccine-induced immunity may impact subsequent de novo responses to drifted epitopes in SARS-CoV-2 variants, but this has been difficult to quantify due to the challenges in recruiting unvaccinated control groups whose first exposure to SARS-CoV-2 is a primary infection. Through local, statewide, and national SARS-CoV-2 testing programs, we were able to recruit cohorts of individuals who had recovered from either primary or post-vaccination infections by either the Delta or Omicron BA.1 variants. Regardless of variant, we observed greater Spike-specific and neutralizing antibody responses in post-vaccination infections than in those who were infected without prior vaccination. Through analysis of variant-specific memory B cells as markers of de novo responses, we observed that Delta and Omicron BA.1 infections led to a marked shift in immunodominance in which some drifted epitopes elicited minimal responses, even in primary infections. Prior immunity through vaccination had a small negative impact on these de novo responses, but this did not correlate with cross-reactive memory B cells, arguing against competitive inhibition of naïve B cells. We conclude that dampened de novo B cell responses against drifted epitopes are mostly a function of altered immunodominance hierarchies that are apparent even in primary infections, with a more modest contribution from pre-existing immunity, perhaps due to accelerated antigen clearance.
Figures





References
-
- Novel 2019 coronavirus genome (2020). Virological. https://virological.org/t/novel-2019-coronavirus-genome/319.
-
- Tanriover M.D., Doğanay H.L., Akova M., Güner H.R., Azap A., Akhan S., Köse Ş., Erdinç F.Ş., Akalın E.H., Tabak Ö.F., et al. (2021). Efficacy and safety of an inactivated whole-virion SARS-CoV-2 vaccine (CoronaVac): interim results of a double-blind, randomised, placebo-controlled, phase 3 trial in Turkey. Lancet 398, 213–222. 10.1016/S0140-6736(21)01429-X. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
