This is a preprint.
Interferon-α promotes neo-antigen formation and preferential HLA-B-restricted antigen presentation in pancreatic β-cells
- PMID: 37745505
- PMCID: PMC10516036
- DOI: 10.1101/2023.09.15.557918
Interferon-α promotes neo-antigen formation and preferential HLA-B-restricted antigen presentation in pancreatic β-cells
Abstract
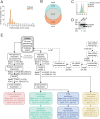
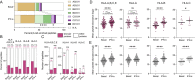
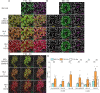
Interferon (IFN)-α is the earliest cytokine signature observed in individuals at risk for type 1 diabetes (T1D), but its effect on the repertoire of HLA Class I (HLA-I)-bound peptides presented by pancreatic β-cells is unknown. Using immunopeptidomics, we characterized the peptide/HLA-I presentation in in-vitro resting and IFN-α-exposed β-cells. IFN-α increased HLA-I expression and peptide presentation, including neo-sequences derived from alternative mRNA splicing, post-translational modifications - notably glutathionylation - and protein cis-splicing. This antigenic landscape relied on processing by both the constitutive and immune proteasome. The resting β-cell immunopeptidome was dominated by HLA-A-restricted ligands. However, IFN-α only marginally upregulated HLA-A and largely favored HLA-B, translating into a major increase in HLA-B-restricted peptides and into an increased activation of HLA-B-restricted vs. HLA-A-restricted CD8+ T-cells. A preferential HLA-B hyper-expression was also observed in the islets of T1D vs. non-diabetic donors, and we identified islet-infiltrating CD8+ T-cells from T1D donors reactive to HLA-B-restricted granule peptides. Thus, the inflammatory milieu of insulitis may skew the autoimmune response toward epitopes presented by HLA-B, hence recruiting a distinct T-cell repertoire that may be relevant to T1D pathogenesis.
Conflict of interest statement
Competing interests. No potential conflicts of interest relevant to this article were reported.
Figures



Similar articles
-
Interferon-α promotes HLA-B-restricted presentation of conventional and alternative antigens in human pancreatic β-cells.Nat Commun. 2025 Jan 17;16(1):765. doi: 10.1038/s41467-025-55908-9. Nat Commun. 2025. PMID: 39824805 Free PMC article.
-
Low HLA binding of diabetes-associated CD8+ T-cell epitopes is increased by post translational modifications.BMC Immunol. 2018 Mar 21;19(1):12. doi: 10.1186/s12865-018-0250-3. BMC Immunol. 2018. PMID: 29562882 Free PMC article.
-
Conventional and Neo-antigenic Peptides Presented by β Cells Are Targeted by Circulating Naïve CD8+ T Cells in Type 1 Diabetic and Healthy Donors.Cell Metab. 2018 Dec 4;28(6):946-960.e6. doi: 10.1016/j.cmet.2018.07.007. Epub 2018 Aug 2. Cell Metab. 2018. PMID: 30078552
-
Immunogenic self-peptides - the great unknowns in autoimmunity: Identifying T-cell epitopes driving the autoimmune response in autoimmune diseases.Front Immunol. 2023 Jan 9;13:1097871. doi: 10.3389/fimmu.2022.1097871. eCollection 2022. Front Immunol. 2023. PMID: 36700227 Free PMC article. Review.
-
Making Insulin and Staying Out of Autoimmune Trouble: The Beta-Cell Conundrum.Front Immunol. 2021 Mar 29;12:639682. doi: 10.3389/fimmu.2021.639682. eCollection 2021. Front Immunol. 2021. PMID: 33854508 Free PMC article. Review.
References
-
- Le Bon A. et al. Type i interferons potently enhance humoral immunity and can promote isotype switching by stimulating dendritic cells in vivo. Immunity 14, 461–470 (2001). - PubMed
-
- Luft T. et al. Type I IFNs enhance the terminal differentiation of dendritic cells. J Immunol 161, 1947–1953 (1998). - PubMed
-
- Marroqui L. et al. Interferon-alpha mediates human beta cell HLA class I overexpression, endoplasmic reticulum stress and apoptosis, three hallmarks of early human type 1 diabetes. Diabetologia 60, 656–667 (2017). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
