This is a preprint.
Immune Evasion, Infectivity, and Fusogenicity of SARS-CoV-2 Omicron BA.2.86 and FLip Variants
- PMID: 37745517
- PMCID: PMC10515800
- DOI: 10.1101/2023.09.11.557206
Immune Evasion, Infectivity, and Fusogenicity of SARS-CoV-2 Omicron BA.2.86 and FLip Variants
Update in
-
Immune evasion, infectivity, and fusogenicity of SARS-CoV-2 BA.2.86 and FLip variants.Cell. 2024 Feb 1;187(3):585-595.e6. doi: 10.1016/j.cell.2023.12.026. Epub 2024 Jan 8. Cell. 2024. PMID: 38194968 Free PMC article.
Abstract
Evolution of SARS-CoV-2 requires the reassessment of current vaccine measures. Here, we characterized BA.2.86 and the XBB-lineage variant FLip by investigating their neutralization alongside D614G, BA.1, BA.2, BA.4/5, XBB.1.5, and EG.5.1 by sera from 3-dose vaccinated and bivalent vaccinated healthcare workers, XBB.1.5-wave infected first responders, and monoclonal antibody (mAb) S309. We assessed the biology of the variant Spikes by measuring viral infectivity and membrane fusogenicity. BA.2.86 is less immune evasive compared to FLip and other XBB variants, consistent with antigenic distances. Importantly, distinct from XBB variants, mAb S309 was unable to neutralize BA.2.86, likely due to a D339H mutation based on modeling. BA.2.86 had relatively high fusogenicity and infectivity in CaLu-3 cells but low fusion and infectivity in 293T-ACE2 cells compared to some XBB variants, suggesting a potentially differences conformational stability of BA.2.86 Spike. Overall, our study underscores the importance of SARS-CoV-2 variant surveillance and the need for updated COVID-19 vaccines.
Conflict of interest statement
Declaration of interests The authors do not declare any competing interests.
Figures
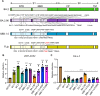

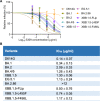
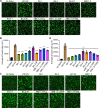



References
-
- Zeng C., Evans J.P., Qu P., Faraone J., Zheng Y.M., Carlin C., Bednash J.S., Zhou T., Lozanski G., Mallampalli R., et al. (2021). Neutralization and Stability of SARS-CoV-2 Omicron Variant. bioRxiv. 10.1101/2021.12.16.472934. - DOI
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
