This is a preprint.
Structural Variation Detection and Association Analysis of Whole-Genome-Sequence Data from 16,905 Alzheimer's Diseases Sequencing Project Subjects
- PMID: 37745545
- PMCID: PMC10516060
- DOI: 10.1101/2023.09.13.23295505
Structural Variation Detection and Association Analysis of Whole-Genome-Sequence Data from 16,905 Alzheimer's Diseases Sequencing Project Subjects
Update in
-
Structural variation detection and association analysis of whole-genome-sequence data from 16,543 Alzheimer's disease sequencing project subjects.Alzheimers Dement. 2025 Jun;21(6):e70277. doi: 10.1002/alz.70277. Alzheimers Dement. 2025. PMID: 40528303 Free PMC article.
Abstract
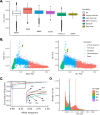
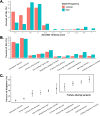
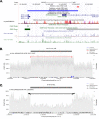
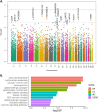
Structural variations (SVs) are important contributors to the genetics of numerous human diseases. However, their role in Alzheimer's disease (AD) remains largely unstudied due to challenges in accurately detecting SVs. Here, we analyzed whole-genome sequencing data from the Alzheimer's Disease Sequencing Project (ADSP, N=16,905 subjects) and identified 400,234 (168,223 high-quality) SVs. We found a significant burden of deletions and duplications in AD cases (OR=1.05, P=0.03), particularly for singletons (OR=1.12, P=0.0002) and homozygous events (OR=1.10, P<0.0004). On AD genes, the ultra-rare SVs, including protein-altering SVs in ABCA7, APP, PLCG2, and SORL1, were associated with AD (SKAT-O P=0.004). Twenty-one SVs are in linkage disequilibrium (LD) with known AD-risk variants, e.g., a deletion (chr2:105731359-105736864) in complete LD (R2=0.99) with rs143080277 (chr2:105749599) in NCK2. We also identified 16 SVs associated with AD and 13 SVs associated with AD-related pathological/cognitive endophenotypes. Our findings demonstrate the broad impact of SVs on AD genetics.
Keywords: Alzheimer’s disease; Copy number variation; Structural variation.
Conflict of interest statement
Competing interests The authors declare that they have no competing interests.
Figures
Similar articles
-
Structural Variation Detection and Association Analysis of Whole-Genome-Sequence Data from 16,905 Alzheimer's Diseases Sequencing Project Subjects.Res Sq [Preprint]. 2023 Oct 5:rs.3.rs-3353179. doi: 10.21203/rs.3.rs-3353179/v1. Res Sq. 2023. Update in: Alzheimers Dement. 2025 Jun;21(6):e70277. doi: 10.1002/alz.70277. PMID: 37886469 Free PMC article. Updated. Preprint.
-
Structural variation detection and association analysis of whole-genome-sequence data from 16,543 Alzheimer's disease sequencing project subjects.Alzheimers Dement. 2025 Jun;21(6):e70277. doi: 10.1002/alz.70277. Alzheimers Dement. 2025. PMID: 40528303 Free PMC article.
-
Vitamin E for Alzheimer's dementia and mild cognitive impairment.Cochrane Database Syst Rev. 2017 Apr 18;4(4):CD002854. doi: 10.1002/14651858.CD002854.pub5. Cochrane Database Syst Rev. 2017. PMID: 28418065 Free PMC article.
-
Vitamin E for Alzheimer's dementia and mild cognitive impairment.Cochrane Database Syst Rev. 2017 Jan 27;1(1):CD002854. doi: 10.1002/14651858.CD002854.pub4. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2017 Apr 18;4:CD002854. doi: 10.1002/14651858.CD002854.pub5. PMID: 28128435 Free PMC article. Updated.
-
Genome structural variants shape adaptive success of an invasive urban malaria vector Anopheles stephensi.bioRxiv [Preprint]. 2024 Jul 30:2024.07.29.605641. doi: 10.1101/2024.07.29.605641. bioRxiv. 2024. Update in: Mol Biol Evol. 2025 Jun 4;42(6):msaf140. doi: 10.1093/molbev/msaf140. PMID: 39211149 Free PMC article. Updated. Preprint.
Cited by
-
The Role of Structural Variants in the Genetic Architecture of Parkinson's Disease.Int J Mol Sci. 2024 Apr 27;25(9):4801. doi: 10.3390/ijms25094801. Int J Mol Sci. 2024. PMID: 38732020 Free PMC article. Review.
References
-
- Gaugler J. et al. 2022 Alzheimer’s disease facts and figures. ALZHEIMERS Dement. 18, 700–789 (2022). - PubMed
-
- Gatz M. et al. Role of genes and environments for explaining Alzheimer disease. Arch. Gen. Psychiatry 63, 168–174 (2006). - PubMed
-
- Farrer L. A. et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease: a meta-analysis. Jama 278, 1349–1356 (1997). - PubMed
Publication types
Grants and funding
- N01 HC085080/HL/NHLBI NIH HHS/United States
- I 904/FWF_/Austrian Science Fund FWF/Austria
- R01 NS080820/NS/NINDS NIH HHS/United States
- P50 NS071674/NS/NINDS NIH HHS/United States
- P20 AG068082/AG/NIA NIH HHS/United States
- P30 AG072975/AG/NIA NIH HHS/United States
- P30 AG066444/AG/NIA NIH HHS/United States
- RC2 HL102419/HL/NHLBI NIH HHS/United States
- P30 AG066507/AG/NIA NIH HHS/United States
- P30 AG072946/AG/NIA NIH HHS/United States
- U01 AG058654/AG/NIA NIH HHS/United States
- U01 HL096812/HL/NHLBI NIH HHS/United States
- U54 AG052427/AG/NIA NIH HHS/United States
- R01 NS017950/NS/NINDS NIH HHS/United States
- P30 AG066518/AG/NIA NIH HHS/United States
- R01 AG019771/AG/NIA NIH HHS/United States
- R01 AG054076/AG/NIA NIH HHS/United States
- RF1 AG074328/AG/NIA NIH HHS/United States
- P20 AG068053/AG/NIA NIH HHS/United States
- R01 AG009029/AG/NIA NIH HHS/United States
- R01 NS069719/NS/NINDS NIH HHS/United States
- R01 AG015928/AG/NIA NIH HHS/United States
- P01 NS026630/NS/NINDS NIH HHS/United States
- P30 AG010133/AG/NIA NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- R01 AG009956/AG/NIA NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- P50 AG016574/AG/NIA NIH HHS/United States
- P30 AG066511/AG/NIA NIH HHS/United States
- N01 HC085082/HL/NHLBI NIH HHS/United States
- P30 AG066512/AG/NIA NIH HHS/United States
- U01 HL096917/HL/NHLBI NIH HHS/United States
- R01 AG019085/AG/NIA NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- P30 AG066515/AG/NIA NIH HHS/United States
- U01 HL130114/HL/NHLBI NIH HHS/United States
- P30 AG062421/AG/NIA NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- N01 HC085083/HL/NHLBI NIH HHS/United States
- P50 NS039764/NS/NINDS NIH HHS/United States
- P30 AG066508/AG/NIA NIH HHS/United States
- U01 HL096902/HL/NHLBI NIH HHS/United States
- U24 AG056270/AG/NIA NIH HHS/United States
- P30 AG072978/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- RC2 HG005605/HG/NHGRI NIH HHS/United States
- P30 AG062429/AG/NIA NIH HHS/United States
- P30 AG066519/AG/NIA NIH HHS/United States
- R01 AG028786/AG/NIA NIH HHS/United States
- R01 AG049607/AG/NIA NIH HHS/United States
- RF1 AG054023/AG/NIA NIH HHS/United States
- P50 AG005136/AG/NIA NIH HHS/United States
- P30 AG072973/AG/NIA NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- P30 AG062422/AG/NIA NIH HHS/United States
- R01 AG079280/AG/NIA NIH HHS/United States
- RF1 AG051504/AG/NIA NIH HHS/United States
- U24 AG041689/AG/NIA NIH HHS/United States
- P30 AG066462/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- P30 AG066530/AG/NIA NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- R01 AG032990/AG/NIA NIH HHS/United States
- N01 HC085079/HL/NHLBI NIH HHS/United States
- U01 HL096814/HL/NHLBI NIH HHS/United States
- P30 AG066509/AG/NIA NIH HHS/United States
- U01 AG006781/AG/NIA NIH HHS/United States
- R01 AG041797/AG/NIA NIH HHS/United States
- K23 AG030944/AG/NIA NIH HHS/United States
- RC4 AG039085/AG/NIA NIH HHS/United States
- P20 AG068077/AG/NIA NIH HHS/United States
- U01 AG062602/AG/NIA NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- R01 AG033040/AG/NIA NIH HHS/United States
- R01 AG021547/AG/NIA NIH HHS/United States
- R01 AG048927/AG/NIA NIH HHS/United States
- R01 AG019757/AG/NIA NIH HHS/United States
- U01 AG046139/AG/NIA NIH HHS/United States
- P30 AG072977/AG/NIA NIH HHS/United States
- R01 AG020098/AG/NIA NIH HHS/United States
- P30 AG062677/AG/NIA NIH HHS/United States
- R01 HL070825/HL/NHLBI NIH HHS/United States
- R01 AG027944/AG/NIA NIH HHS/United States
- P20 AG068024/AG/NIA NIH HHS/United States
- P30 AG072958/AG/NIA NIH HHS/United States
- R01 AG025259/AG/NIA NIH HHS/United States
- U01 HL096899/HL/NHLBI NIH HHS/United States
- P30 AG062715/AG/NIA NIH HHS/United States
- RF1 AG015473/AG/NIA NIH HHS/United States
- P30 AG066506/AG/NIA NIH HHS/United States
- P30 AG066468/AG/NIA NIH HHS/United States
- P30 AG072976/AG/NIA NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- R01 AG011101/AG/NIA NIH HHS/United States
- P30 AG072947/AG/NIA NIH HHS/United States
- P30 AG072931/AG/NIA NIH HHS/United States
- P30 AG072972/AG/NIA NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- P30 AG066514/AG/NIA NIH HHS/United States
- P30 AG072959/AG/NIA NIH HHS/United States
- U19 AG066567/AG/NIA NIH HHS/United States
- N01 HC085081/HL/NHLBI NIH HHS/United States
- P30 AG072979/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
