This is a preprint.
Genomic consequences of isolation and inbreeding in an island dingo population
- PMID: 37745583
- PMCID: PMC10516007
- DOI: 10.1101/2023.09.15.557950
Genomic consequences of isolation and inbreeding in an island dingo population
Update in
-
Genomic Consequences of Isolation and Inbreeding in an Island Dingo Population.Genome Biol Evol. 2024 Jul 3;16(7):evae130. doi: 10.1093/gbe/evae130. Genome Biol Evol. 2024. PMID: 38913571 Free PMC article.
Abstract
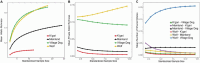
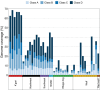
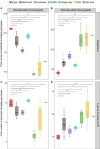
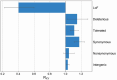
Dingoes come from an ancient canid lineage that originated in East Asia around 8000-11,000 years BP. As Australia's largest terrestrial predator, dingoes play an important ecological role. A small, protected population exists on a world heritage listed offshore island, K'gari (formerly Fraser Island). Concern regarding the persistence of dingoes on K'gari has risen due to their low genetic diversity and elevated inbreeding levels. However, whole-genome sequencing data is lacking from this population. Here, we include five new whole-genome sequences of K'gari dingoes. We analyze a total of 18 whole genome sequences of dingoes sampled from mainland Australia and K'gari to assess the genomic consequences of their demographic histories. Long (>1 Mb) runs of homozygosity (ROH) - indicators of inbreeding - are elevated in all sampled dingoes. However, K'gari dingoes showed significantly higher levels of very long ROH (>5 Mb), providing genomic evidence for small population size, isolation, inbreeding, and a strong founder effect. Our results suggest that, despite current levels of inbreeding, the K'gari population is purging strongly deleterious mutations, which, in the absence of further reductions in population size, may facilitate the persistence of small populations despite low genetic diversity and isolation. However, there may be little to no purging of mildly deleterious alleles, which may have important long-term consequences, and should be considered by conservation and management programs.
Conflict of interest statement
CONFLICTS OF INTEREST KMC is a scientific advisor to the Australian Dingo Foundation, The New Guinea Singing Dog Conservation Society and The New Guinea Highland Wild Dog Foundation. KMC and LB are members of the IUCN Canid Specialist Group Dingo Working Group.
Figures

Similar articles
-
Genomic Consequences of Isolation and Inbreeding in an Island Dingo Population.Genome Biol Evol. 2024 Jul 3;16(7):evae130. doi: 10.1093/gbe/evae130. Genome Biol Evol. 2024. PMID: 38913571 Free PMC article.
-
Conservation concerns associated with low genetic diversity for K'gari-Fraser Island dingoes.Sci Rep. 2021 May 4;11(1):9503. doi: 10.1038/s41598-021-89056-z. Sci Rep. 2021. PMID: 33947920 Free PMC article.
-
Efficacy of Management Efforts to Reduce Food-Related Dingo-Human Interactions and Conflict on K'gari (Fraser Island), Australia.Animals (Basel). 2023 Jan 5;13(2):204. doi: 10.3390/ani13020204. Animals (Basel). 2023. PMID: 36670744 Free PMC article.
-
Insects for breakfast and whales for dinner: the diet and body condition of dingoes on Fraser Island (K'gari).Sci Rep. 2016 Mar 24;6:23469. doi: 10.1038/srep23469. Sci Rep. 2016. PMID: 27009879 Free PMC article.
-
Runs of homozygosity: current knowledge and applications in livestock.Anim Genet. 2017 Jun;48(3):255-271. doi: 10.1111/age.12526. Epub 2016 Dec 1. Anim Genet. 2017. PMID: 27910110 Review.
References
-
- Allen B. 2023. Genetic health and status of K’gari wongari (Fraser Island dingoes). Queensland Government - Department of Environment and Science [Internet]. Available from: https://parks.des.qld.gov.au/_data/assets/pdf_file/0022/319414/genetic-h...
-
- Allen BL, Higginbottom K, Bracks JH, Davies N, Baxter GS. 2015. Balancing dingo conservation with human safety on Fraser Island: the numerical and demographic effects of humane destruction of dingoes. Australas. J. Environ. Manag. 22:197–215.
-
- Behrendorff L. 2021. Best-practice dingo management: six lessons from K’gari (Fraser Island). Aust. Zool. 41:521–533.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
