Insights into the physiological, molecular, and genetic regulators of albinism in Camellia sinensis leaves
- PMID: 37745858
- PMCID: PMC10516542
- DOI: 10.3389/fgene.2023.1219335
Insights into the physiological, molecular, and genetic regulators of albinism in Camellia sinensis leaves
Abstract
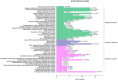
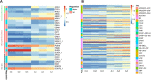
Introduction: Yanling Yinbiancha, a cultivar of Camellia sinensis (L.) O. Kuntze, is an evergreen woody perennial with characteristic albino leaves. A mutant variant with green leaves on branches has been recently identified. The molecular mechanisms underlying this color variation remain unknown. Methods: We aimed to utilize omics tools to decipher the molecular basis for this color variation, with the ultimate goal of enhancing existing germplasm and utilizing it in future breeding programs. Results and discussion: Albinotic leaves exhibited significant chloroplast degeneration and reduced carotenoid accumulation. Transcriptomic and metabolomic analysis of the two variants revealed 1,412 differentially expressed genes and 127 differentially accumulated metabolites (DAMs). Enrichment analysis for DEGs suggested significant enrichment of pathways involved in the biosynthesis of anthocyanins, porphyrin, chlorophyll, and carotenoids. To further narrow down the causal variation for albinotic leaves, we performed a conjoint analysis of metabolome and transcriptome and identified putative candidate genes responsible for albinism in C. sinensis leaves. 12, 7, and 28 DEGs were significantly associated with photosynthesis, porphyrin/chlorophyll metabolism, and flavonoid metabolism, respectively. Chlorophyllase 2, Chlorophyll a-Binding Protein 4A, Chlorophyll a-Binding Protein 24, Stay Green Regulator, Photosystem II Cytochrome b559 subunit beta along with transcription factors AP2, bZIP, MYB, and WRKY were identified as a potential regulator of albinism in Yanling Yinbiancha. Moreover, we identified Anthocyanidin reductase and Arabidopsis Response Regulator 1 as DEGs influencing flavonoid accumulation in albino leaves. Identification of genes related to albinism in C. sinensis may facilitate genetic modification or development of molecular markers, potentially enhancing cultivation efficiency and expanding the germplasm for utilization in breeding programs.
Keywords: albinism; carotenoids; chlorophyll; flavonoids; leaves; pigmentation.
Copyright © 2023 Zhao, Yang, Cheng, Liu, Yang and Liu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Ahmed S., Stepp J. R., Orians C., Griffin T., Matyas C., Robbat A., et al. (2014). Effects of extreme climate events on tea (Camellia sinensis) functional quality validate indigenous farmer knowledge and sensory preferences in tropical China. PloS one 9, e109126. 10.1371/journal.pone.0109126 - DOI - PMC - PubMed
-
- Barry C. S. (2009). The stay-green revolution: recent progress in deciphering the mechanisms of chlorophyll degradation in higher plants. Plant Sci. 176, 325–333. 10.1016/j.plantsci.2008.12.013 - DOI
-
- Bo L., Zhang F., Zhang L.-W., Huang J.-F., Zhi-Feng J., Gupta D. (2012). Comprehensive suitability evaluation of tea crops using GIS and a modified land ecological suitability evaluation model. Pedosphere 22, 122–130. 10.1016/s1002-0160(11)60198-7 - DOI
LinkOut - more resources
Full Text Sources
Research Materials

