Screening and validation of novel serum panel of microRNA in stratification of prostate cancer
- PMID: 37745909
- PMCID: PMC10513910
- DOI: 10.1016/j.prnil.2023.06.002
Screening and validation of novel serum panel of microRNA in stratification of prostate cancer
Erratum in
-
Corrigendum to "Screening and validation of novel serum panel of microRNA in stratification of prostate cancer" [Prostate Int 11 (2023) 150-158].Prostate Int. 2024 Sep;12(3):178. doi: 10.1016/j.prnil.2024.01.001. Epub 2024 Jan 20. Prostate Int. 2024. PMID: 39816934 Free PMC article.
Abstract
Background: Owing to the heterogeneous nature of prostate cancer (PCa) and errors in the characterization of the disease, researchers have been trying to unveil molecular biomarkers like microRNA (miRNA) as diagnostic markers. The purpose of our study is to demonstrate the precision of a panel of miRNAs as biomarkers with diagnostic potential for risk stratification.
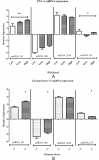
Materials and methods: The present study demonstrates the comparative expression profiles of miRNA-141,-1290,-100, and -335 in both tissue and serum, including Benign Prostate Hyperplasia (BPH) and PCa, with healthy volunteers. Firstly, we demonstrate the expression of all miRNAs in the discovery cohort, including metastasis and benign tissue, and later validate their non-invasive diagnostic potential in BPH and PCa with healthy volunteers. MiRNA was isolated from tissue and serum to be quantified by RT-PCR and analyzed for biomarker potential by receiver operating characteristic (ROC) curve analysis, followed by targetome analysis of each miRNA.
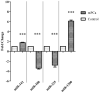
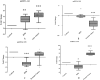
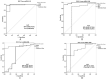
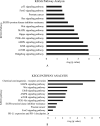
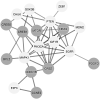
Results: Among the non-invasive miRNA assessed, it was seen that miRNA 141 (P = 0.0003) and miRNA 1290 (P < 0.0001) are oncogenic with significantly higher expression, while miRNA 100 (P = 0.0002) and miRNA 335 are tumor suppressor, in PCa as compared to controls. While for BPH, miRNA 141 (P = 0.003) and miRNA 335 (P = 0.0002) were found to be significantly oncogenic and tumor suppressors, respectively. The analysis of the ROC curve of panel miRNAs (miRNA-141,-1290, and -100) portrayed a significant area under the curve with greater sensitivity and specificity. Moreover, in-silico prediction of their respective targetomes represents their extensive involvement in PCa progression and various other cascades that aid in PCa networks.
Conclusions: To the best of our knowledge, we are going to report for the first time this panel of miRNA that can be used to accurately and efficiently diagnose BPH and PCa patients from healthy males.
Keywords: Benign prostatic hyperplasia; Biomarker; Diagnosis; MicroRNA; Non-invasive; Prostate cancer.
© 2023 The Asian Pacific Prostate Society. Published by Elsevier B.V.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Heidenreich A., Bellmunt J., Bolla M., Joniau S., Mason M., Matveev V., et al. EAU guidelines on prostate cancer. Part 1: screening, diagnosis, and treatment of clinically localised disease. Eur Urol. 2011 Jan;59(1):61–71. doi: 10.1016/j.eururo.2013.09.046. Epub 2013 Oct 6. PMID: 24207135. - DOI - PubMed
-
- Canto E.I., Singh H., Shariat S.F., Lamb D.J., Mikolajczyk S.D., Linton H.J., et al. Serum BPSA outperforms both total PSA and free PSA as a predictor of prostatic enlargement in men without prostate cancer. Urology. 2004 May;63(5):905–910. doi: 10.1016/j.urology.2003.12.037. PMID: 15134977. - DOI - PubMed
-
- Cho H., Oh C.K., Cha J., Chung J.I., Byun S.S., Hong S.K., et al. Association of serum prostate-specific antigen (PSA) level and circulating tumor cell-based PSA mRNA in prostate cancer. Prostate Int. 2022 Mar;10(1):14–20. doi: 10.1016/j.prnil.2022.01.002. Epub 2022 Jan 10. PMID: 35229001; PMCID: PMC8844604. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
