Basidiomycota Fungi and ROS: Genomic Perspective on Key Enzymes Involved in Generation and Mitigation of Reactive Oxygen Species
- PMID: 37746164
- PMCID: PMC10512322
- DOI: 10.3389/ffunb.2022.837605
Basidiomycota Fungi and ROS: Genomic Perspective on Key Enzymes Involved in Generation and Mitigation of Reactive Oxygen Species
Abstract
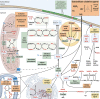
Our review includes a genomic survey of a multitude of reactive oxygen species (ROS) related intra- and extracellular enzymes and proteins among fungi of Basidiomycota, following their taxonomic classification within the systematic classes and orders, and focusing on different fungal lifestyles (saprobic, symbiotic, pathogenic). Intra- and extracellular ROS metabolism-involved enzymes (49 different protein families, summing 4170 protein models) were searched as protein encoding genes among 63 genomes selected according to current taxonomy. Extracellular and intracellular ROS metabolism and mechanisms in Basidiomycota are illustrated in detail. In brief, it may be concluded that differences between the set of extracellular enzymes activated by ROS, especially by H2O2, and involved in generation of H2O2, follow the differences in fungal lifestyles. The wood and plant biomass degrading white-rot fungi and the litter-decomposing species of Agaricomycetes contain the highest counts for genes encoding various extracellular peroxidases, mono- and peroxygenases, and oxidases. These findings further confirm the necessity of the multigene families of various extracellular oxidoreductases for efficient and complete degradation of wood lignocelluloses by fungi. High variations in the sizes of the extracellular ROS-involved gene families were found, however, among species with mycorrhizal symbiotic lifestyle. In addition, there are some differences among the sets of intracellular thiol-mediation involving proteins, and existence of enzyme mechanisms for quenching of intracellular H2O2 and ROS. In animal- and plant-pathogenic species, extracellular ROS enzymes are absent or rare. In these fungi, intracellular peroxidases are seemingly in minor role than in the independent saprobic, filamentous species of Basidiomycota. Noteworthy is that our genomic survey and review of the literature point to that there are differences both in generation of extracellular ROS as well as in mechanisms of response to oxidative stress and mitigation of ROS between fungi of Basidiomycota and Ascomycota.
Keywords: Basidiomycota; CAZy AA auxiliary enzymes; GMC oxidoreductases; NADPH oxidase (NOX); catalase (CAT); reactive oxygen species (ROS); superoxide dismutase; thioredoxin (TRX) family proteins.
Copyright © 2022 Mattila, Österman-Udd, Mali and Lundell.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling editor LN declared a past collaboration with the authors TL.
Figures



Similar articles
-
A survey of genes encoding H2O2-producing GMC oxidoreductases in 10 Polyporales genomes.Mycologia. 2015 Nov-Dec;107(6):1105-19. doi: 10.3852/15-027. Epub 2015 Aug 21. Mycologia. 2015. PMID: 26297778
-
Gene Regulation Shifts Shed Light on Fungal Adaption in Plant Biomass Decomposers.mBio. 2019 Nov 19;10(6):e02176-19. doi: 10.1128/mBio.02176-19. mBio. 2019. PMID: 31744914 Free PMC article.
-
Genomic Analysis Enlightens Agaricales Lifestyle Evolution and Increasing Peroxidase Diversity.Mol Biol Evol. 2021 Apr 13;38(4):1428-1446. doi: 10.1093/molbev/msaa301. Mol Biol Evol. 2021. PMID: 33211093 Free PMC article.
-
Linking the Metabolic Activity of Plastic-Degrading Fungi to Their Taxonomy and Evolution.J Fungi (Basel). 2025 May 15;11(5):378. doi: 10.3390/jof11050378. J Fungi (Basel). 2025. PMID: 40422712 Free PMC article. Review.
-
Lignin-modifying enzymes in filamentous basidiomycetes--ecological, functional and phylogenetic review.J Basic Microbiol. 2010 Feb;50(1):5-20. doi: 10.1002/jobm.200900338. J Basic Microbiol. 2010. PMID: 20175122 Review.
Cited by
-
Nitrogen limitation causes a seismic shift in redox state and phosphorylation of proteins implicated in carbon flux and lipidome remodeling in Rhodotorula toruloides.Biotechnol Biofuels Bioprod. 2025 Jul 21;18(1):80. doi: 10.1186/s13068-025-02657-y. Biotechnol Biofuels Bioprod. 2025. PMID: 40691609 Free PMC article.
-
Methionine oxidation of carbohydrate-active enzymes during white-rot wood decay.Appl Environ Microbiol. 2024 Mar 20;90(3):e0193123. doi: 10.1128/aem.01931-23. Epub 2024 Feb 20. Appl Environ Microbiol. 2024. PMID: 38376171 Free PMC article.
-
Hydrated lime promoted the polysaccharide content and affected the transcriptomes of Lentinula edodes during brown film formation.Front Microbiol. 2023 Dec 4;14:1290180. doi: 10.3389/fmicb.2023.1290180. eCollection 2023. Front Microbiol. 2023. PMID: 38111638 Free PMC article.
-
Substrate specificity mapping of fungal CAZy AA3_2 oxidoreductases.Biotechnol Biofuels Bioprod. 2024 Mar 27;17(1):47. doi: 10.1186/s13068-024-02491-8. Biotechnol Biofuels Bioprod. 2024. PMID: 38539167 Free PMC article.
-
Oxidative stress and culture atmosphere effects on bioactive compounds and laccase activity in the white rot fungus Phlebia radiata on birch wood substrate.Curr Res Microb Sci. 2024 Sep 19;7:100280. doi: 10.1016/j.crmicr.2024.100280. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39398196 Free PMC article.
References
-
- Alfaro M., Castanera R., Lavín J. L., Grigoriev I. V., Oguiza J. A., Ramírez L., et al. . (2016). Comparative and transcriptional analysis of the predicted secretome in the lignocellulose-degrading basidiomycete fungus Pleurotus ostreatus. Environ. Microbiol. 18, 4710–4726. 10.1111/1462-2920.13360 - DOI - PubMed
-
- Arantes V., Goodell B. (2014). Current understanding of brown-rot fungal biodegradation mechanisms: a review. ACS Symposium Ser. 1158, 3–21. 10.1021/bk-2014-1158.ch001 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

