Genomic characterization of polyextremotolerant black yeasts isolated from food and food production environments
- PMID: 37746166
- PMCID: PMC10512282
- DOI: 10.3389/ffunb.2022.928622
Genomic characterization of polyextremotolerant black yeasts isolated from food and food production environments
Abstract
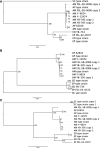
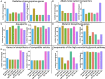
Black yeasts have been isolated from acidic, low water activity, and thermally processed foods as well as from surfaces in food manufacturing plants. The genomic basis for their relative tolerance to food-relevant environmental stresses has not been well defined. In this study, we performed whole genome sequencing (WGS) on seven black yeast strains including Aureobasidium (n=5) and Exophiala (n=2) which were isolated from food or food production environments. These strains were previously characterized for their tolerance to heat, hyperosmotic pressure, high pressure processing, hypochlorite sanitizers, and ultraviolet light. Based on the WGS data, three of the strains previously identified as A. pullulans were reassigned as A. melanogenum. Both haploid and diploid A. melanogenum strains were identified in this collection. Single-locus phylogenies based on beta tubulin, RNA polymerase II, or translation elongation factor protein sequences were compared to the phylogeny produced through SNP analysis, revealing that duplication of the fungal genome in diploid strains complicates the use of single-locus phylogenetics. There was not a strong association between phylogeny and either environmental source or stress tolerance phenotype, nor were trends in the copy numbers of stress-related genes associated with extremotolerance within this collection. While there were obvious differences between the genera, the heterogenous distribution of stress tolerance phenotypes and genotypes suggests that food-relevant black yeasts may be ubiquitous rather than specialists associated with particular ecological niches. However, further evaluation of additional strains and the potential impact of gene sequence modification is necessary to confirm these findings.
Keywords: Aureobasidium; Exophiala; food mycology; genome duplication; single-gene phylogeny.
Copyright © 2022 Cai and Snyder.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





Similar articles
-
Thermoresistance in Black Yeasts Is Associated with Halosensitivity and High Pressure Processing Tolerance but Not with UV Tolerance or Sanitizer Tolerance.J Food Prot. 2022 Feb 1;85(2):203-212. doi: 10.4315/JFP-21-314. J Food Prot. 2022. PMID: 34614188
-
Clonality, inbreeding, and hybridization in two extremotolerant black yeasts.Gigascience. 2022 Oct 6;11:giac095. doi: 10.1093/gigascience/giac095. Gigascience. 2022. PMID: 36200832 Free PMC article.
-
Redefinition of Aureobasidium pullulans and its varieties.Stud Mycol. 2008;61:21-38. doi: 10.3114/sim.2008.61.02. Stud Mycol. 2008. PMID: 19287524 Free PMC article.
-
Pervasive Listeria monocytogenes Is Common in the Norwegian Food System and Is Associated with Increased Prevalence of Stress Survival and Resistance Determinants.Appl Environ Microbiol. 2022 Sep 22;88(18):e0086122. doi: 10.1128/aem.00861-22. Epub 2022 Aug 25. Appl Environ Microbiol. 2022. PMID: 36005805 Free PMC article. Review.
-
Black yeasts in the omics era: Achievements and challenges.Med Mycol. 2018 Apr 1;56(suppl_1):32-41. doi: 10.1093/mmy/myx129. Med Mycol. 2018. PMID: 29538737 Review.
Cited by
-
Microbial food spoilage: impact, causative agents and control strategies.Nat Rev Microbiol. 2024 Sep;22(9):528-542. doi: 10.1038/s41579-024-01037-x. Epub 2024 Apr 3. Nat Rev Microbiol. 2024. PMID: 38570695 Review.
-
Genomic deletions in Aureobasidium pullulans by an AMA1 plasmid for gRNA and CRISPR/Cas9 expression.Fungal Biol Biotechnol. 2024 Jun 1;11(1):6. doi: 10.1186/s40694-024-00175-4. Fungal Biol Biotechnol. 2024. PMID: 38824542 Free PMC article.
References
-
- Andrews S. (2010) FastQC: A quality control tool for high throughput sequence data [Online]. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
-
- Bell A. A., Wheeler M. H. (1986). Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 24, 411–451. doi: 10.1146/annurev.py.24.090186.002211 - DOI
LinkOut - more resources
Full Text Sources

