Antifungal Exposure and Resistance Development: Defining Minimal Selective Antifungal Concentrations and Testing Methodologies
- PMID: 37746188
- PMCID: PMC10512330
- DOI: 10.3389/ffunb.2022.918717
Antifungal Exposure and Resistance Development: Defining Minimal Selective Antifungal Concentrations and Testing Methodologies
Abstract
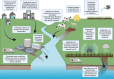
This scoping review aims to summarise the current understanding of selection for antifungal resistance (AFR) and to compare and contrast this with selection for antibacterial resistance, which has received more research attention. AFR is an emerging global threat to human health, associated with high mortality rates, absence of effective surveillance systems and with few alternative treatment options available. Clinical AFR is well documented, with additional settings increasingly being recognised to play a role in the evolution and spread of AFR. The environment, for example, harbours diverse fungal communities that are regularly exposed to antifungal micropollutants, potentially increasing AFR selection risk. The direct application of effect concentrations of azole fungicides to agricultural crops and the incomplete removal of pharmaceutical antifungals in wastewater treatment systems are of particular concern. Currently, environmental risk assessment (ERA) guidelines do not require assessment of antifungal agents in terms of their ability to drive AFR development, and there are no established experimental tools to determine antifungal selective concentrations. Without data to interpret the selective risk of antifungals, our ability to effectively inform safe environmental thresholds is severely limited. In this review, potential methods to generate antifungal selective concentration data are proposed, informed by approaches used to determine antibacterial minimal selective concentrations. Such data can be considered in the development of regulatory guidelines that aim to reduce selection for AFR.
Keywords: antifungal resistance; antifungals; antimicrobial resistance; experimental evolution; fungi; minimal selective concentration; selection.
Copyright © 2022 Stevenson, Gaze, Gow, Hart, Schmidt, Usher, Warris, Wilkinson and Murray.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Arendrup M. C., Meletiadis J., Mouton J. W., Guinea J., Cuenca-Estrella M., Lagrou K., et al. . (2016). EUCAST Technical Note on Isavuconazole Breakpoints for Aspergillus, Itraconazole Breakpoints for Candida and Updates for the Antifungal Susceptibility Testing Method Documents. Clin. Microbiol. Infect. 22 (6), 571 e571–574. doi: 10.1016/j.cmi.2016.01.017 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

