Discovery of a gene cluster for the biosynthesis of novel cyclic peptide compound, KK-1, in Curvularia clavata
- PMID: 37746209
- PMCID: PMC10512319
- DOI: 10.3389/ffunb.2022.1081179
Discovery of a gene cluster for the biosynthesis of novel cyclic peptide compound, KK-1, in Curvularia clavata
Abstract
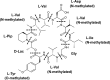
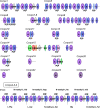
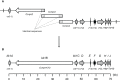
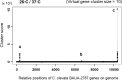
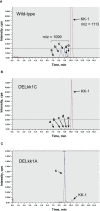
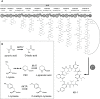
KK-1, a cyclic depsipeptide with 10 residues produced by a filamentous fungus Curvularia clavata BAUA-2787, is a promising pesticide active compound with high activity against many plant pathogens, especially Botrytis cinerea. As a first step toward the future mass production of KK-1 through synthetic biological approaches, we aimed to identify the genes responsible for the KK-1 biosynthesis. To achieve this, we conducted whole genome sequencing and transcriptome analysis of C. clavata BAUA-2787 to predict the KK-1 biosynthetic gene cluster. We then generated the overexpression and deletion mutants for each cluster gene using our originally developed transformation system for this fungus, and analyzed the KK-1 production and the cluster gene expression levels to confirm their involvement in KK-1 biosynthesis. As a result of these, a region of approximately 71 kb was found, containing 10 open reading frames, which were co-induced during KK-1 production, as a biosynthetic gene cluster. These include kk1B, which encodes nonribosomal peptide synthetase with a domain structure that is consistent with the structural features of KK-1, and kk1F, which encodes a transcription factor. The overexpression of kk1F increased the expression of the entire cluster genes and, consequently, improved KK-1 production, whereas its deletion decreased the expression of the entire cluster genes and almost eliminated KK-1 production, demonstrating that the protein encoded by kk1F regulates the expressions of the other nine cluster genes cooperatively as the pathway-specific transcription factor. Furthermore, the deletion of each cluster gene caused a reduction in KK-1 productivity, indicating that each gene is involved in KK-1 production. The genes kk1A, kk1D, kk1H, and kk1I, which showed a significant decrease in KK-1 productivity due to deletion, were presumed to be directly involved in KK-1 structure formation, including the biosynthesis of the constituent residues. kk1C, kk1E, kk1G, and kk1J, which maintained a certain level of KK-1 productivity despite deletion, were possibly involved in promoting or assisting KK-1 production, such as extracellular transportation and the removal of aberrant units incorporated into the peptide chain.
Keywords: Curvularia clavata; antifungal agent; biosynthetic gene cluster; genome sequence; grey mold; natural fungicide; non-ribosomal peptide; pesticide.
Copyright © 2023 Yamaguchi, Fujioka, Yoshimi, Kumagai, Umemura, Abe, Machida and Kawai.
Conflict of interest statement
Author SY, TF, and KK are employed by Kumiai Chemical Industry Co., Ltd. and TK is employed by Fermlab Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Heterologous Production of a Novel Cyclic Peptide Compound, KK-1, in Aspergillus oryzae.Front Microbiol. 2018 Apr 9;9:690. doi: 10.3389/fmicb.2018.00690. eCollection 2018. Front Microbiol. 2018. PMID: 29686660 Free PMC article.
-
Characterization of a gene cluster responsible for the biosynthesis of anticancer agent FK228 in Chromobacterium violaceum No. 968.Appl Environ Microbiol. 2007 Jun;73(11):3460-9. doi: 10.1128/AEM.01751-06. Epub 2007 Mar 30. Appl Environ Microbiol. 2007. PMID: 17400765 Free PMC article.
-
Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster.Appl Environ Microbiol. 2010 Dec;76(24):8143-9. doi: 10.1128/AEM.00683-10. Epub 2010 Oct 15. Appl Environ Microbiol. 2010. PMID: 20952652 Free PMC article.
-
Functional characterization and manipulation of the apicidin biosynthetic pathway in Fusarium semitectum.Mol Microbiol. 2010 Apr;76(2):456-66. doi: 10.1111/j.1365-2958.2010.07109.x. Epub 2010 Mar 10. Mol Microbiol. 2010. PMID: 20233305
-
Nonribosomal peptides in fungal cell factories: from genome mining to optimized heterologous production.Biotechnol Adv. 2019 Dec;37(8):107449. doi: 10.1016/j.biotechadv.2019.107449. Epub 2019 Sep 10. Biotechnol Adv. 2019. PMID: 31518630 Review.
Cited by
-
Regulation and induction of fungal secondary metabolites: a comprehensive review.Arch Microbiol. 2025 Jul 1;207(8):189. doi: 10.1007/s00203-025-04386-0. Arch Microbiol. 2025. PMID: 40590991 Review.
-
Natural Products That Contain Higher Homologated Amino Acids.Chembiochem. 2024 May 2;25(9):e202300822. doi: 10.1002/cbic.202300822. Epub 2024 Apr 9. Chembiochem. 2024. PMID: 38487927 Free PMC article. Review.
References
-
- Brzozowski L., Mazourek M. (2018). A sustainable agricultural future relies on the transition to organic agroecological pest management. Sustainability. 10:2023. doi: 10.3390/su10062023 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

