Small NRPS-like enzymes in Aspergillus sections Flavi and Circumdati selectively form substituted pyrazinone metabolites
- PMID: 37746228
- PMCID: PMC10512218
- DOI: 10.3389/ffunb.2022.1029195
Small NRPS-like enzymes in Aspergillus sections Flavi and Circumdati selectively form substituted pyrazinone metabolites
Abstract
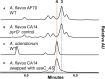
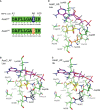
Aspergillus fungi produce mycotoxins that are detrimental to human and animal health. Two sections of aspergilli are of particular importance to cereal food crops such as corn and barley. Aspergillus section Flavi species like A. flavus and A. parasiticus produce aflatoxins, while section Circumdati species like A. ochraceus and A. sclerotiorum produce ochratoxin A. Mitigating these toxins in food and feed is a critical and ongoing worldwide effort. We have previously investigated biosynthetic gene clusters in Aspergillus flavus that are linked to fungal virulence in corn. We found that one such cluster, asa, is responsible for the production of aspergillic acid, an iron-binding, hydroxamic acid-containing pyrazinone metabolite. Furthermore, we found that the asa gene cluster is present in many other aflatoxin- and ochratoxin-producing aspergilli. The core gene in the asa cluster encodes the small nonribosomal peptide synthetase-like (NRPS-like) protein AsaC. We have swapped the asaC ortholog from A. sclerotiorum into A. flavus, replacing its native copy, and have also cloned both asaC orthologs into Saccharomyces cerevisiae. We show that AsaC orthologs in section Flavi and section Circumdati, while only containing adenylation-thiolation-reductase (ATR) domains, can selectively biosynthesize distinct pyrazinone natural products: deoxyaspergillic acid and flavacol, respectively. Because pyrazinone natural products and the gene clusters responsible for their production are implicated in a variety of important microbe-host interactions, uncovering the function and selectivity of the enzymes involved could lead to strategies that ultimately benefit human health.
Keywords: ATR; aflatoxin; aspergillic acid; biosynthesis; flavacol; mycotoxin; ochratoxin.
Copyright © 2022 Lebar, Mack, Carter-Wientjes, Wei, Mattison and Cary.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Identification and functional analysis of the aspergillic acid gene cluster in Aspergillus flavus.Fungal Genet Biol. 2018 Jul;116:14-23. doi: 10.1016/j.fgb.2018.04.009. Epub 2018 Apr 16. Fungal Genet Biol. 2018. PMID: 29674152
-
Simultaneous detection of mycotoxigenic Aspergillus species of sections Circumdati and Flavi using multiplex digital PCR.Lett Appl Microbiol. 2023 Dec 7;76(12):ovad142. doi: 10.1093/lambio/ovad142. Lett Appl Microbiol. 2023. PMID: 38111225
-
Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B1, sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus rambellii sp. nov.Syst Appl Microbiol. 2005 Jul;28(5):442-53. doi: 10.1016/j.syapm.2005.02.012. Syst Appl Microbiol. 2005. PMID: 16094871
-
Aspergillus Species and Their Associated Mycotoxins.Methods Mol Biol. 2017;1542:33-49. doi: 10.1007/978-1-4939-6707-0_3. Methods Mol Biol. 2017. PMID: 27924530 Review.
-
The Secondary Metabolites and Biosynthetic Diversity From Aspergillus ochraceus.Front Chem. 2022 Aug 25;10:938626. doi: 10.3389/fchem.2022.938626. eCollection 2022. Front Chem. 2022. PMID: 36092677 Free PMC article. Review.
Cited by
-
Next-generation AMA1-based plasmids for enhanced heterologous expression in filamentous fungi.Microb Biotechnol. 2024 Sep;17(9):e70010. doi: 10.1111/1751-7915.70010. Microb Biotechnol. 2024. PMID: 39276061 Free PMC article.
-
Allantofuranone Biosynthesis and Precursor-Directed Mutasynthesis of Hydroxylated Analogues.J Nat Prod. 2025 May 23;88(5):1191-1200. doi: 10.1021/acs.jnatprod.5c00197. Epub 2025 Apr 18. J Nat Prod. 2025. PMID: 40247749 Free PMC article.
-
ZfpA-Dependent Quorum Sensing Shifts in Morphology and Secondary Metabolism in Aspergillus flavus.Environ Microbiol. 2025 Apr;27(4):e70100. doi: 10.1111/1462-2920.70100. Environ Microbiol. 2025. PMID: 40262766 Free PMC article.
-
Identification and heterologous expression of an NRPS biosynthetic gene cluster responsible for the production of the pyrazinones Ichizinone A, B and C.Microb Cell Fact. 2025 Jun 7;24(1):131. doi: 10.1186/s12934-025-02753-6. Microb Cell Fact. 2025. PMID: 40481494 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

