Mechanistic modeling explains the production dynamics of recombinant adeno-associated virus with the baculovirus expression vector system
- PMID: 37746245
- PMCID: PMC10512016
- DOI: 10.1016/j.omtm.2023.05.019
Mechanistic modeling explains the production dynamics of recombinant adeno-associated virus with the baculovirus expression vector system
Abstract
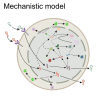
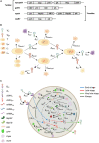
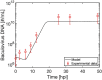
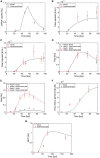
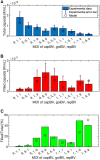
Current manufacturing processes for recombinant adeno-associated viruses (rAAVs) have less-than-desired yields and produce significant amounts of empty capsids. The increasing demand and the high cost of goods for rAAV-based gene therapies motivate development of more efficient manufacturing processes. Recently, the US Food and Drug Administration (FDA) approved the first rAAV-based gene therapy product manufactured in the baculovirus expression vector system (BEVS), a technology that demonstrated production of high titers of full capsids. This work presents a first mechanistic model describing the key extracellular and intracellular phenomena occurring during baculovirus infection and rAAV maturation in the BEVS. The model predictions are successfully validated for in-house and literature experimental measurements of the vector genome and of structural and non-structural proteins collected during rAAV manufacturing in the BEVS with the TwoBac and ThreeBac constructs. A model-based analysis of the process is carried out to identify the bottlenecks that limit full capsid formation. Vector genome amplification is found to be the limiting step for rAAV production in Sf9 cells using either the TwoBac or ThreeBac system. In turn, vector genome amplification is hindered by limiting Rep78 levels. Transgene and non-essential baculovirus protein expression in the insect cell during rAAV manufacturing also negatively influences the rAAV production yields.
Keywords: AAV-based gene therapy; baculovirus expression vector system; biomanufacturing; gene therapy; mechanistic modeling; recombinant adeno-associated virus; viral vector; viral vector manufacturing.
© 2023 The Author(s).
Conflict of interest statement
R.M.K. is an inventor on patents related to recombinant AAV technology and owns equity in gene therapy-related companies. Portions of the recombinant AAV technology studied in this report are covered by United States and European patents assigned to the Secretary of the U.S. Department of Health and Human Services. A fraction of the licensing fees and royalty payments made to the National Institutes of Health is distributed to the inventors (R.M.K.) in accordance with U.S. Government and National Institutes of Health policy.
Figures


Similar articles
-
Monobac System-A Single Baculovirus for the Production of rAAV.Microorganisms. 2021 Aug 24;9(9):1799. doi: 10.3390/microorganisms9091799. Microorganisms. 2021. PMID: 34576695 Free PMC article.
-
Impact of dual-baculovirus infection on the Sf9 insect cell transcriptome during rAAV production using single-cell RNA-seq.Biotechnol Bioeng. 2023 Sep;120(9):2588-2600. doi: 10.1002/bit.28377. Epub 2023 Apr 7. Biotechnol Bioeng. 2023. PMID: 36919374
-
Establishment of a Recombinant AAV2/HBoV1 Vector Production System in Insect Cells.Genes (Basel). 2020 Apr 17;11(4):439. doi: 10.3390/genes11040439. Genes (Basel). 2020. PMID: 32316599 Free PMC article.
-
Advancements in molecular design and bioprocessing of recombinant adeno-associated virus gene delivery vectors using the insect-cell baculovirus expression platform.Biotechnol J. 2021 Apr;16(4):e2000021. doi: 10.1002/biot.202000021. Epub 2021 Jan 25. Biotechnol J. 2021. PMID: 33277815 Review.
-
Molecular design for recombinant adeno-associated virus (rAAV) vector production.Appl Microbiol Biotechnol. 2018 Feb;102(3):1045-1054. doi: 10.1007/s00253-017-8670-1. Epub 2017 Dec 4. Appl Microbiol Biotechnol. 2018. PMID: 29204900 Free PMC article. Review.
Cited by
-
Enhancement of exogenous protein stability in AcMNPV by overexpressing lef5 gene during passaging.Appl Microbiol Biotechnol. 2025 Mar 24;109(1):73. doi: 10.1007/s00253-025-13451-z. Appl Microbiol Biotechnol. 2025. PMID: 40126614 Free PMC article.
-
Systematic comparison of rAAV vectors manufactured using large-scale suspension cultures of Sf9 and HEK293 cells.Mol Ther. 2024 Jan 3;32(1):74-83. doi: 10.1016/j.ymthe.2023.11.022. Epub 2023 Nov 20. Mol Ther. 2024. PMID: 37990495 Free PMC article.
-
A clinician's guide to AAV production - How manufacturing platforms shape vector properties.Med Genet. 2025 Jul 17;37(3):169-177. doi: 10.1515/medgen-2025-2024. eCollection 2025 Jul. Med Genet. 2025. PMID: 40687886 Free PMC article.
References
-
- U.S. National Library of Medicine . 2022. ClinicalTrials.gov.https://www.clinicaltrials.gov
-
- Evaluate Ltd . 2022. Evaluate Pharma 2022.https://www.evaluate.com/thought-leadership/pharma/world-preview-2022-re...
-
- Rininger J., Fennell A., Schoukroun-Barnes L., Peterson C., Speidel J. 2019. Capacity Analysis for Viral Vector Manufacturing: Is There Enough?https://bioprocessintl.com/manufacturing/emerging-therapeutics-manufactu...
LinkOut - more resources
Full Text Sources

