Application of Protein-Protein Interaction Network Analysis in Order to Identify Cervical Cancer miRNA and mRNA Biomarkers
- PMID: 37746664
- PMCID: PMC10513823
- DOI: 10.1155/2023/6626279
Application of Protein-Protein Interaction Network Analysis in Order to Identify Cervical Cancer miRNA and mRNA Biomarkers
Abstract
Cervical cancer (CC) is one of the world's most common and severe cancers. This cancer includes two histological types: squamous cell carcinoma (SCC) and adenocarcinoma (ADC). The current study aims at identifying novel potential candidate mRNA and miRNA biomarkers for SCC based on a protein-protein interaction (PPI) and miRNA-mRNA network analysis. The current project utilized a transcriptome profile for normal and SCC samples. First, the PPI network was constructed for the 1335 DEGs, and then, a significant gene module was extracted from the PPI network. Next, a list of miRNAs targeting module's genes was collected from the experimentally validated databases, and a miRNA-mRNA regulatory network was formed. After network analysis, four driver genes were selected from the module's genes including MCM2, MCM10, POLA1, and TONSL and introduced as potential candidate biomarkers for SCC. In addition, two hub miRNAs, including miR-193b-3p and miR-615-3p, were selected from the miRNA-mRNA regulatory network and reported as possible candidate biomarkers. In summary, six potential candidate RNA-based biomarkers consist of four genes containing MCM2, MCM10, POLA1, and TONSL, and two miRNAs containing miR-193b-3p and miR-615-3p are opposed as potential candidate biomarkers for CC.
Copyright © 2023 Parinaz Tabrizi-Nezhadi et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures

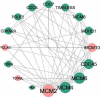
References
-
- Shaw P., Senthilnathan R., Sankar S., et al. A clinical investigation on the theragnostic effect of MicroRNA biomarkers for survival outcome in cervical cancer: a prisma-P compliant protocol for systematic review and comprehensive meta-analysis. Genes . 2022;13(3):p. 463. doi: 10.3390/genes13030463. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

