Expansion of Pathogenic Cardiac Macrophages in Immune Checkpoint Inhibitor Myocarditis
- PMID: 37746718
- PMCID: PMC11323830
- DOI: 10.1161/CIRCULATIONAHA.122.062551
Expansion of Pathogenic Cardiac Macrophages in Immune Checkpoint Inhibitor Myocarditis
Abstract
Background: Immune checkpoint inhibitors (ICIs), antibodies targeting PD-1 (programmed cell death protein 1)/PD-L1 (programmed death-ligand 1) or CTLA4 (cytotoxic T-lymphocyte-associated protein 4), have revolutionized cancer management but are associated with devastating immune-related adverse events including myocarditis. The main risk factor for ICI myocarditis is the use of combination PD-1 and CTLA4 inhibition. ICI myocarditis is often fulminant and is pathologically characterized by myocardial infiltration of T lymphocytes and macrophages. Although much has been learned about the role of T-cells in ICI myocarditis, little is understood about the identity, transcriptional diversity, and functions of infiltrating macrophages.
Methods: We used an established murine ICI myocarditis model (Ctla4+/-Pdcd1-/- mice) to explore the cardiac immune landscape using single-cell RNA-sequencing, immunostaining, flow cytometry, in situ RNA hybridization, molecular imaging, and antibody neutralization studies.
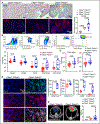
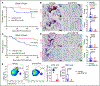
Results: We observed marked increases in CCR2 (C-C chemokine receptor type 2)+ monocyte-derived macrophages and CD8+ T-cells in this model. The macrophage compartment was heterogeneous and displayed marked enrichment in an inflammatory CCR2+ subpopulation highly expressing Cxcl9 (chemokine [C-X-C motif] ligand 9), Cxcl10 (chemokine [C-X-C motif] ligand 10), Gbp2b (interferon-induced guanylate-binding protein 2b), and Fcgr4 (Fc receptor, IgG, low affinity IV) that originated from CCR2+ monocytes. It is important that a similar macrophage population expressing CXCL9, CXCL10, and CD16α (human homologue of mouse FcgR4) was expanded in patients with ICI myocarditis. In silico prediction of cell-cell communication suggested interactions between T-cells and Cxcl9+Cxcl10+ macrophages via IFN-γ (interferon gamma) and CXCR3 (CXC chemokine receptor 3) signaling pathways. Depleting CD8+ T-cells or macrophages and blockade of IFN-γ signaling blunted the expansion of Cxcl9+Cxcl10+ macrophages in the heart and attenuated myocarditis, suggesting that this interaction was necessary for disease pathogenesis.
Conclusions: These data demonstrate that ICI myocarditis is associated with the expansion of a specific population of IFN-γ-induced inflammatory macrophages and suggest the possibility that IFN-γ blockade may be considered as a treatment option for this devastating condition.
Keywords: CXCL9 chemokine; IFN-gamma; T-cells; cytotoxic T lymphocyte-associated antigen 4-immunoglobulin; macrophages; myocarditis; programmed cell death protein 1.
Conflict of interest statement
Figures






Update of
-
Expansion of Disease Specific Cardiac Macrophages in Immune Checkpoint Inhibitor Myocarditis.bioRxiv [Preprint]. 2023 Apr 29:2023.04.28.538426. doi: 10.1101/2023.04.28.538426. bioRxiv. 2023. Update in: Circulation. 2024 Jan 2;149(1):48-66. doi: 10.1161/CIRCULATIONAHA.122.062551. PMID: 37162929 Free PMC article. Updated. Preprint.
Comment in
-
T Cells and Macrophages Drive Pathogenesis of Immune Checkpoint Inhibitor Myocarditis.Circulation. 2024 Jan 2;149(1):67-69. doi: 10.1161/CIRCULATIONAHA.123.067189. Epub 2023 Dec 28. Circulation. 2024. PMID: 38153995 No abstract available.
References
MeSH terms
Substances
Grants and funding
- R35 HL145212/HL/NHLBI NIH HHS/United States
- R01 HL151685/HL/NHLBI NIH HHS/United States
- R01 HL150891/HL/NHLBI NIH HHS/United States
- P30 AR073752/AR/NIAMS NIH HHS/United States
- P41 EB025815/EB/NIBIB NIH HHS/United States
- R01 HL156021/HL/NHLBI NIH HHS/United States
- R01 HL141466/HL/NHLBI NIH HHS/United States
- R01 HL131908/HL/NHLBI NIH HHS/United States
- R01 HL153436/HL/NHLBI NIH HHS/United States
- R01 HL151078/HL/NHLBI NIH HHS/United States
- R01 HL139714/HL/NHLBI NIH HHS/United States
- R01 HL138466/HL/NHLBI NIH HHS/United States
- R01 HL155990/HL/NHLBI NIH HHS/United States
- R35 HL161185/HL/NHLBI NIH HHS/United States
- R01 HL160688/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

