Broad and potent neutralizing antibodies are elicited in vaccinated individuals following Delta/BA.1 breakthrough infection
- PMID: 37747187
- PMCID: PMC10653880
- DOI: 10.1128/mbio.01206-23
Broad and potent neutralizing antibodies are elicited in vaccinated individuals following Delta/BA.1 breakthrough infection
Abstract
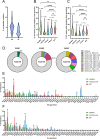
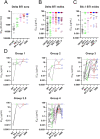
With the emergence of SARS-CoV-2 viral variants, there has been an increase in infections in vaccinated individuals. Here, we isolated monoclonal antibodies (mAbs) from individuals experiencing a breakthrough infection (Delta or BA.1) to determine how exposure to a heterologous Spike broadens the neutralizing antibody response at the monoclonal level. All mAbs isolated had reactivity to the Spike of the vaccine and infection variant. While many mAbs showed reduced neutralization of current circulating variants, we identified mAbs with broad and potent neutralization of BA.2.75.2, XBB, XBB.1.5, and BQ.1.1 indicating the presence of conserved epitopes on Spike. These results indicate that variant-based vaccine boosters have the potential to broaden the vaccine response.
Keywords: SARS-CoV-2; immunization; infectious disease; monoclonal antibodies; neutralizing antibodies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang M-L, Jerome KR, Bloom JD, Greninger AL. 2020. Neutralizing antibodies correlate with protection from SARS-CoV-2 in humans during a fishery vessel outbreak with a high attack rate. J Clin Microbiol 58:e02107-20. doi:10.1128/JCM.02107-20 - DOI - PMC - PubMed
-
- Lustig Y, Sapir E, Regev-Yochay G, Cohen C, Fluss R, Olmer L, Indenbaum V, Mandelboim M, Doolman R, Amit S, Mendelson E, Ziv A, Huppert A, Rubin C, Freedman L, Kreiss Y. 2021. BNT162b2 COVID-19 vaccine and correlates of humoral immune responses and dynamics: a prospective, single-centre, longitudinal cohort study in health-care workers. Lancet Respir Med 9:999–1009. doi:10.1016/S2213-2600(21)00220-4 - DOI - PMC - PubMed
-
- Graham C, Seow J, Huettner I, Khan H, Kouphou N, Acors S, Winstone H, Pickering S, Galao RP, Dupont L, Lista MJ, Jimenez-Guardeño JM, Laing AG, Wu Y, Joseph M, Muir L, van Gils MJ, Ng WM, Duyvesteyn HME, Zhao Y, Bowden TA, Shankar-Hari M, Rosa A, Cherepanov P, McCoy LE, Hayday AC, Neil SJD, Malim MH, Doores KJ. 2021. Neutralization potency of monoclonal antibodies recognizing dominant and subdominant epitopes on SARS-CoV-2 spike is impacted by the B.1.1.7 variant. Immunity 54:1276–1289. doi:10.1016/j.immuni.2021.03.023 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

