Proteotoxic stresses stimulate dissociation of UBL4A from the tail-anchored protein recognition complex
- PMID: 37747814
- PMCID: PMC10586765
- DOI: 10.1042/BCJ20230267
Proteotoxic stresses stimulate dissociation of UBL4A from the tail-anchored protein recognition complex
Abstract
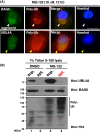
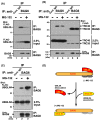
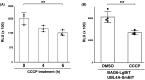
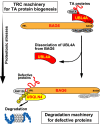
Inclusion body formation is associated with cytotoxicity in a number of neurodegenerative diseases. However, the molecular basis of the toxicity caused by the accumulation of aggregation-prone proteins remains controversial. In this study, we found that disease-associated inclusions induced by elongated polyglutamine chains disrupt the complex formation of BAG6 with UBL4A, a mammalian homologue of yeast Get5. UBL4A also dissociated from BAG6 in response to proteotoxic stresses such as proteasomal inhibition and mitochondrial depolarization. These findings imply that the cytotoxicity of pathological protein aggregates might be attributed in part to disruption of the BAG6-UBL4A complex that is required for the biogenesis of tail-anchored proteins.
Keywords: BAG6; UBL4A; polyglutamine disease; proteasome; protein quality control; tail-anchored protein.
© 2023 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures



Similar articles
-
Structure of a BAG6 (Bcl-2-associated athanogene 6)-Ubl4a (ubiquitin-like protein 4a) complex reveals a novel binding interface that functions in tail-anchored protein biogenesis.J Biol Chem. 2015 Apr 10;290(15):9387-98. doi: 10.1074/jbc.M114.631804. Epub 2015 Feb 20. J Biol Chem. 2015. PMID: 25713138 Free PMC article.
-
Nuclear BAG6-UBL4A-GET4 complex mediates DNA damage signaling and cell death.J Biol Chem. 2013 Jul 12;288(28):20547-57. doi: 10.1074/jbc.M112.443416. Epub 2013 May 30. J Biol Chem. 2013. PMID: 23723067 Free PMC article.
-
Bag6 complex contains a minimal tail-anchor-targeting module and a mock BAG domain.Proc Natl Acad Sci U S A. 2015 Jan 6;112(1):106-11. doi: 10.1073/pnas.1402745112. Epub 2014 Dec 22. Proc Natl Acad Sci U S A. 2015. PMID: 25535373 Free PMC article.
-
The roles of cytosolic quality control proteins, SGTA and the BAG6 complex, in disease.Adv Protein Chem Struct Biol. 2019;114:265-313. doi: 10.1016/bs.apcsb.2018.11.002. Epub 2018 Dec 18. Adv Protein Chem Struct Biol. 2019. PMID: 30635083 Free PMC article. Review.
-
BAG6/BAT3: emerging roles in quality control for nascent polypeptides.J Biochem. 2013 Feb;153(2):147-60. doi: 10.1093/jb/mvs149. Epub 2012 Dec 28. J Biochem. 2013. PMID: 23275523 Review.
References
-
- Igarashi, S., Koide, R., Shimohata, T., Yamada, M., Hayashi, Y., Takano, H.et al. (1998) Suppression of aggregate formation and apoptosis by transglutaminase inhibitors in cells expressing truncated DRPLA protein with an expanded polyglutamine stretch. Nat. Genet. 18, 111–117 10.1038/ng0298-111 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

