ADAR-mediated regulation of PQM-1 expression in neurons impacts gene expression throughout C. elegans and regulates survival from hypoxia
- PMID: 37747897
- PMCID: PMC10553819
- DOI: 10.1371/journal.pbio.3002150
ADAR-mediated regulation of PQM-1 expression in neurons impacts gene expression throughout C. elegans and regulates survival from hypoxia
Abstract
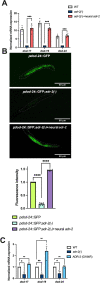
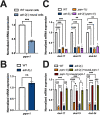
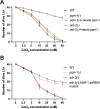
The ability to alter gene expression programs in response to changes in environmental conditions is central to the ability of an organism to thrive. For most organisms, the nervous system serves as the master regulator in communicating information about the animal's surroundings to other tissues. The information relay centers on signaling pathways that cue transcription factors in a given cell type to execute a specific gene expression program, but also provide a means to signal between tissues. The transcription factor PQM-1 is an important mediator of the insulin signaling pathway contributing to longevity and the stress response as well as impacting survival from hypoxia. Herein, we reveal a novel mechanism for regulating PQM-1 expression specifically in neural cells of larval animals. Our studies reveal that the RNA-binding protein (RBP), ADR-1, binds to pqm-1 mRNA in neural cells. This binding is regulated by the presence of a second RBP, ADR-2, which when absent leads to reduced expression of both pqm-1 and downstream PQM-1 activated genes. Interestingly, we find that neural pqm-1 expression is sufficient to impact gene expression throughout the animal and affect survival from hypoxia, phenotypes that we also observe in adr mutant animals. Together, these studies reveal an important posttranscriptional gene regulatory mechanism in Caenorhabditis elegans that allows the nervous system to sense and respond to environmental conditions to promote organismal survival from hypoxia.
Copyright: © 2023 Mahapatra et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
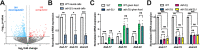

Update of
-
ADARs employ a neural-specific mechanism to regulate PQM-1 expression and survival from hypoxia.bioRxiv [Preprint]. 2023 May 5:2023.05.05.539519. doi: 10.1101/2023.05.05.539519. bioRxiv. 2023. Update in: PLoS Biol. 2023 Sep 25;21(9):e3002150. doi: 10.1371/journal.pbio.3002150. PMID: 37205482 Free PMC article. Updated. Preprint.
References
-
- Padilla PA, Nystul TG, Zager RA, Johnson AC, Roth MB. Dephosphorylation of cell cycle-regulated proteins correlates with anoxia-induced suspended animation in Caenorhabditis elegans. Mol Biol Cell. 2002;13(5):1473–1483. doi: 10.1091/mbc.01-12-0594 ; PubMed Central PMCID: PMC111120. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

