Distinguishing features of long COVID identified through immune profiling
- PMID: 37748514
- PMCID: PMC10620090
- DOI: 10.1038/s41586-023-06651-y
Distinguishing features of long COVID identified through immune profiling
Abstract
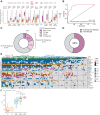
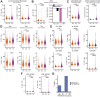
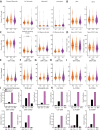
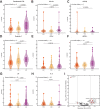
Post-acute infection syndromes may develop after acute viral disease1. Infection with SARS-CoV-2 can result in the development of a post-acute infection syndrome known as long COVID. Individuals with long COVID frequently report unremitting fatigue, post-exertional malaise, and a variety of cognitive and autonomic dysfunctions2-4. However, the biological processes that are associated with the development and persistence of these symptoms are unclear. Here 275 individuals with or without long COVID were enrolled in a cross-sectional study that included multidimensional immune phenotyping and unbiased machine learning methods to identify biological features associated with long COVID. Marked differences were noted in circulating myeloid and lymphocyte populations relative to the matched controls, as well as evidence of exaggerated humoral responses directed against SARS-CoV-2 among participants with long COVID. Furthermore, higher antibody responses directed against non-SARS-CoV-2 viral pathogens were observed among individuals with long COVID, particularly Epstein-Barr virus. Levels of soluble immune mediators and hormones varied among groups, with cortisol levels being lower among participants with long COVID. Integration of immune phenotyping data into unbiased machine learning models identified the key features that are most strongly associated with long COVID status. Collectively, these findings may help to guide future studies into the pathobiology of long COVID and help with developing relevant biomarkers.
© 2023. The Author(s).
Conflict of interest statement
In the past three years, H.M.K. received expenses and/or personal fees from UnitedHealth, Element Science, Eyedentifeye and F-Prime; he is a co-founder of Refactor Health and HugoHealth; and is associated with contracts, through Yale New Haven Hospital, from the Centers for Medicare & Medicaid Services and through Yale University from the Food and Drug Administration, Johnson & Johnson, Google and Pfizer. N.K. is a scientific founder at Thyron; served as a consultant to Boehringer Ingelheim, Pliant, Astra Zeneca, RohBar, Veracyte, Galapagos, Fibrogen and Thyron over the past 3 years; reports equity in Pliant and Thyron; and acknowledges grants from Veracyte, Boehringer Ingelheim and BMS. A.I. co-founded and consults for RIGImmune, Xanadu Bio and PanV; consults for Paratus Sciences and InvisiShield Technologies; and is a member of the Board of Directors of Roche Holding. A.M.R. and Y.D. are listed as inventors on a patent describing the REAP technology. A.M.R. is the founder and director of Seranova Bio. A.M.R. and Y.D. hold equity in Seranova Bio. The other authors declare no competing interests.
Figures










Update of
-
Distinguishing features of Long COVID identified through immune profiling.medRxiv [Preprint]. 2022 Aug 10:2022.08.09.22278592. doi: 10.1101/2022.08.09.22278592. medRxiv. 2022. Update in: Nature. 2023 Nov;623(7985):139-148. doi: 10.1038/s41586-023-06651-y. PMID: 35982667 Free PMC article. Updated. Preprint.
Comment in
-
The distinctive immune features of long COVID.Nat Rev Immunol. 2023 Nov;23(11):703. doi: 10.1038/s41577-023-00958-7. Nat Rev Immunol. 2023. PMID: 37816952 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

