Two domains of Tim50 coordinate translocation of proteins across the two mitochondrial membranes
- PMID: 37748811
- PMCID: PMC10520260
- DOI: 10.26508/lsa.202302122
Two domains of Tim50 coordinate translocation of proteins across the two mitochondrial membranes
Abstract
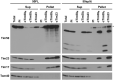
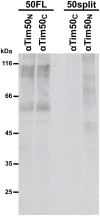
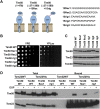
Hundreds of mitochondrial proteins with N-terminal presequences are translocated across the outer and inner mitochondrial membranes via the TOM and TIM23 complexes, respectively. How translocation of proteins across two mitochondrial membranes is coordinated is largely unknown. Here, we show that the two domains of Tim50 in the intermembrane space, named core and PBD, both have essential roles in this process. Building upon the surprising observation that the two domains of Tim50 can complement each other in trans, we establish that the core domain contains the main presequence-binding site and serves as the main recruitment point to the TIM23 complex. On the other hand, the PBD plays, directly or indirectly, a critical role in cooperation of the TOM and TIM23 complexes and supports the receptor function of Tim50. Thus, the two domains of Tim50 both have essential but distinct roles and together coordinate translocation of proteins across two mitochondrial membranes.
© 2023 Genge et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures







Similar articles
-
Tim50 is a subunit of the TIM23 complex that links protein translocation across the outer and inner mitochondrial membranes.Cell. 2002 Nov 15;111(4):519-28. doi: 10.1016/s0092-8674(02)01053-x. Cell. 2002. PMID: 12437925
-
Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria.Mol Biol Cell. 2009 Mar;20(5):1400-7. doi: 10.1091/mbc.e08-09-0934. Epub 2009 Jan 14. Mol Biol Cell. 2009. PMID: 19144822 Free PMC article.
-
Structural basis for the function of Tim50 in the mitochondrial presequence translocase.J Mol Biol. 2011 Aug 19;411(3):513-9. doi: 10.1016/j.jmb.2011.06.020. Epub 2011 Jun 17. J Mol Biol. 2011. PMID: 21704637 Free PMC article.
-
From TOM to the TIM23 complex - handing over of a precursor.Biol Chem. 2020 May 26;401(6-7):709-721. doi: 10.1515/hsz-2020-0101. Biol Chem. 2020. PMID: 32074073 Review.
-
Unlocking the presequence import pathway.Trends Cell Biol. 2015 May;25(5):265-75. doi: 10.1016/j.tcb.2014.12.001. Epub 2014 Dec 23. Trends Cell Biol. 2015. PMID: 25542066 Review.
Cited by
-
Dbi1 is an oxidoreductase and an assembly chaperone for mitochondrial inner membrane proteins.EMBO Rep. 2025 Feb;26(4):911-928. doi: 10.1038/s44319-024-00349-6. Epub 2025 Jan 3. EMBO Rep. 2025. PMID: 39753782 Free PMC article.
-
Molecular machineries and pathways of mitochondrial protein transport.Nat Rev Mol Cell Biol. 2025 Jul 3. doi: 10.1038/s41580-025-00865-w. Online ahead of print. Nat Rev Mol Cell Biol. 2025. PMID: 40610778 Review.
References
-
- Ahting U, Floss T, Uez N, Schneider-Lohmar I, Becker L, Kling E, Iuso A, Bender A, de Angelis MH, Gailus-Durner V, et al. (2009) Neurological phenotype and reduced lifespan in heterozygous Tim23 knockout mice, the first mouse model of defective mitochondrial import. Biochim Biophys Acta 1787: 371–376. 10.1016/j.bbabio.2008.12.001 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
