Identification of a miRNA multi-targeting therapeutic strategy in glioblastoma
- PMID: 37749143
- PMCID: PMC10519979
- DOI: 10.1038/s41419-023-06117-z
Identification of a miRNA multi-targeting therapeutic strategy in glioblastoma
Abstract
Glioblastoma (GBM) is a deadly and the most common primary brain tumor in adults. Due to their regulation of a high number of mRNA transcripts, microRNAs (miRNAs) are key molecules in the control of biological processes and are thereby promising therapeutic targets for GBM patients. In this regard, we recently reported miRNAs as strong modulators of GBM aggressiveness. Here, using an integrative and comprehensive analysis of the TCGA database and the transcriptome of GBM biopsies, we identified three critical and clinically relevant miRNAs for GBM, miR-17-3p, miR-222, and miR-340. In addition, we showed that the combinatorial modulation of three of these miRNAs efficiently inhibited several biological processes in patient-derived GBM cells of all these three GBM subtypes (Mesenchymal, Proneural, Classical), induced cell death, and delayed tumor growth in a mouse tumor model. Finally, in a doxycycline-inducible model, we observed a significant inhibition of GBM stem cell viability and a significant delay of orthotopic tumor growth. Collectively, our results reveal, for the first time, the potential of miR-17-3p, miR-222 and miR-340 multi-targeting as a promising therapeutic strategy for GBM patients.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
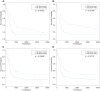



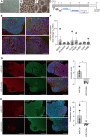
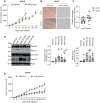
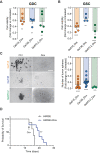
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

