Genomic characterization of rare earth binding by Shewanella oneidensis
- PMID: 37749198
- PMCID: PMC10520059
- DOI: 10.1038/s41598-023-42742-6
Genomic characterization of rare earth binding by Shewanella oneidensis
Abstract
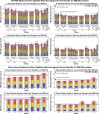
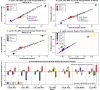
Rare earth elements (REE) are essential ingredients of sustainable energy technologies, but separation of individual REE is one of the hardest problems in chemistry today. Biosorption, where molecules adsorb to the surface of biological materials, offers a sustainable alternative to environmentally harmful solvent extractions currently used for separation of rare earth elements (REE). The REE-biosorption capability of some microorganisms allows for REE separations that, under specialized conditions, are already competitive with solvent extractions, suggesting that genetic engineering could allow it to leapfrog existing technologies. To identify targets for genomic improvement we screened 3,373 mutants from the whole genome knockout collection of the known REE-biosorbing microorganism Shewanella oneidensis MR-1. We found 130 genes that increased biosorption of the middle REE europium, and 112 that reduced it. We verified biosorption changes from the screen for a mixed solution of three REE (La, Eu, Yb) using Inductively Coupled Plasma Mass Spectrometry (ICP-MS) in solution conditions with a range of ionic strengths and REE concentrations. We identified 18 gene ontologies and 13 gene operons that make up key systems that affect biosorption. We found, among other things, that disruptions of a key regulatory component of the arc system (hptA), which regulates cellular response to anoxic environments and polysaccharide biosynthesis related genes (wbpQ, wbnJ, SO_3183) consistently increase biosorption across all our solution conditions. Our largest total biosorption change comes from our SO_4685, a capsular polysaccharide (CPS) synthesis gene, disruption of which results in an up to 79% increase in biosorption; and nusA, a transcriptional termination/anti-termination protein, disruption of which results in an up to 35% decrease in biosorption. Knockouts of glnA, pyrD, and SO_3183 produce small but significant increases (≈ 1%) in relative biosorption affinity for ytterbium over lanthanum in multiple solution conditions tested, while many other genes we explored have more complex binding affinity changes. Modeling suggests that while these changes to lanthanide biosorption selectivity are small, they could already reduce the length of repeated enrichment process by up to 27%. This broad exploratory study begins to elucidate how genetics affect REE-biosorption by S. oneidensis, suggests new areas of investigation for better mechanistic understanding of the membrane chemistry involved in REE binding, and offer potential targets for improving biosorption and separation of REE by genetic engineering.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors are pursuing patent protection for engineered organisms using knowledge gathered in this work (US provisional patent Application no. 63/405353). A.M.S. and S.M. are co-founders of, and B.B. is a contributor, and uncompensated advisor to, REEgen, Inc., which is developing genetically engineered microbes for REE bio-mining. The remaining authors have competing interests to declare.
Figures


References
-
- Dent PC. Rare earth elements and permanent magnets. J. Appl. Phys. 2012;111:07A721. doi: 10.1063/1.3676616. - DOI
-
- Lucas J, Lucas P, Le Mercier T, Rollat A, Davenport W. Rare Earths: Science, Technology, Production and Use. Elsevier Inc.; 2014.
-
- Norman AF, Prangnell PB, McEwen RS. The solidification behaviour of dilute aluminium–scandium alloys. Acta Mater. 1998;46:5715–5732. doi: 10.1016/s1359-6454(98)00257-2. - DOI
-
- Adesina O, Anzai IA, Avalos JL, Barstow B. Embracing biological solutions to the sustainable energy challenge. Chemistry. 2017;2:20–51. doi: 10.1016/j.chempr.2016.12.009. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources

