Iron drives anabolic metabolism through active histone demethylation and mTORC1
- PMID: 37749225
- PMCID: PMC11407783
- DOI: 10.1038/s41556-023-01225-6
Iron drives anabolic metabolism through active histone demethylation and mTORC1
Abstract
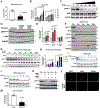
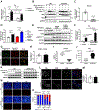
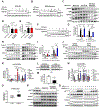
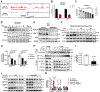
All eukaryotic cells require a minimal iron threshold to sustain anabolic metabolism. However, the mechanisms by which cells sense iron to regulate anabolic processes are unclear. Here we report a previously undescribed eukaryotic pathway for iron sensing in which molecular iron is required to sustain active histone demethylation and maintain the expression of critical components of the pro-anabolic mTORC1 pathway. Specifically, we identify the iron-binding histone-demethylase KDM3B as an intrinsic iron sensor that regulates mTORC1 activity by demethylating H3K9me2 at enhancers of a high-affinity leucine transporter, LAT3, and RPTOR. By directly suppressing leucine availability and RAPTOR levels, iron deficiency supersedes other nutrient inputs into mTORC1. This process occurs in vivo and is not an indirect effect by canonical iron-utilizing pathways. Because ancestral eukaryotes share homologues of KDMs and mTORC1 core components, this pathway probably pre-dated the emergence of the other kingdom-specific nutrient sensors for mTORC1.
© 2023. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing Interests:
The authors declare no competing interests.
Figures




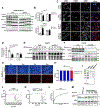

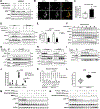
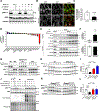

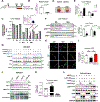

Comment in
-
A new pathway links iron sensing with histone demethylation and regulation of mTORC1 activity.Nat Rev Nephrol. 2023 Dec;19(12):753. doi: 10.1038/s41581-023-00785-y. Nat Rev Nephrol. 2023. PMID: 37884788 No abstract available.
References
Methods-Only References
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL140973/HL/NHLBI NIH HHS/United States
- P41 GM135018/GM/NIGMS NIH HHS/United States
- S10 OD011996/OD/NIH HHS/United States
- R01 GM038784/GM/NIGMS NIH HHS/United States
- P41 GM108569/GM/NIGMS NIH HHS/United States
- P30 CA060553/CA/NCI NIH HHS/United States
- T32 GM008449/GM/NIGMS NIH HHS/United States
- U54 CA193419/CA/NCI NIH HHS/United States
- R01 HL140927/HL/NHLBI NIH HHS/United States
- R01 HL155953/HL/NHLBI NIH HHS/United States
- R01 HL138982/HL/NHLBI NIH HHS/United States
- R01 HL127646/HL/NHLBI NIH HHS/United States
- T32 DK007169/DK/NIDDK NIH HHS/United States
- F30 DK109608/DK/NIDDK NIH HHS/United States
- S10 OD026814/OD/NIH HHS/United States
- P30 DK114857/DK/NIDDK NIH HHS/United States
- S10 OD025194/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

