APOE4 impairs the microglial response in Alzheimer's disease by inducing TGFβ-mediated checkpoints
- PMID: 37749326
- PMCID: PMC10863749
- DOI: 10.1038/s41590-023-01627-6
APOE4 impairs the microglial response in Alzheimer's disease by inducing TGFβ-mediated checkpoints
Abstract
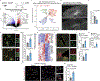
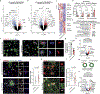
The APOE4 allele is the strongest genetic risk factor for late-onset Alzheimer's disease (AD). The contribution of microglial APOE4 to AD pathogenesis is unknown, although APOE has the most enriched gene expression in neurodegenerative microglia (MGnD). Here, we show in mice and humans a negative role of microglial APOE4 in the induction of the MGnD response to neurodegeneration. Deletion of microglial APOE4 restores the MGnD phenotype associated with neuroprotection in P301S tau transgenic mice and decreases pathology in APP/PS1 mice. MGnD-astrocyte cross-talk associated with β-amyloid (Aβ) plaque encapsulation and clearance are mediated via LGALS3 signaling following microglial APOE4 deletion. In the brains of AD donors carrying the APOE4 allele, we found a sex-dependent reciprocal induction of AD risk factors associated with suppression of MGnD genes in females, including LGALS3, compared to individuals homozygous for the APOE3 allele. Mechanistically, APOE4-mediated induction of ITGB8-transforming growth factor-β (TGFβ) signaling impairs the MGnD response via upregulation of microglial homeostatic checkpoints, including Inpp5d, in mice. Deletion of Inpp5d in microglia restores MGnD-astrocyte cross-talk and facilitates plaque clearance in APP/PS1 mice. We identify the microglial APOE4-ITGB8-TGFβ pathway as a negative regulator of microglial response to AD pathology, and restoring the MGnD phenotype via blocking ITGB8-TGFβ signaling provides a promising therapeutic intervention for AD.
© 2023. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Figures












References
-
- Keren-Shaul H et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290 e1217 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM132668/GM/NIGMS NIH HHS/United States
- P30 DK040561/DK/NIDDK NIH HHS/United States
- RF1 NS090934/NS/NINDS NIH HHS/United States
- R21 NS101673/NS/NINDS NIH HHS/United States
- R01 AG075509/AG/NIA NIH HHS/United States
- K12 EY016335/EY/NEI NIH HHS/United States
- R01 EY027921/EY/NEI NIH HHS/United States
- R01 AG054672/AG/NIA NIH HHS/United States
- K08 EY030160/EY/NEI NIH HHS/United States
- AARF-21-846786/ALZ/Alzheimer's Association/United States
- R21 NS104609/NS/NINDS NIH HHS/United States
- R01 AG051812/AG/NIA NIH HHS/United States
- R01 NS088137/NS/NINDS NIH HHS/United States
- R21 AG076982/AG/NIA NIH HHS/United States
- R01 AG080992/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

