Interaction between estrogen receptor-α and PNPLA3 p.I148M variant drives fatty liver disease susceptibility in women
- PMID: 37749332
- PMCID: PMC10579099
- DOI: 10.1038/s41591-023-02553-8
Interaction between estrogen receptor-α and PNPLA3 p.I148M variant drives fatty liver disease susceptibility in women
Erratum in
-
Author Correction: Interaction between estrogen receptor-α and PNPLA3 p.I148M variant drives fatty liver disease susceptibility in women.Nat Med. 2024 Apr;30(4):1212. doi: 10.1038/s41591-024-02817-x. Nat Med. 2024. PMID: 38267549 Free PMC article. No abstract available.
Abstract
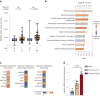
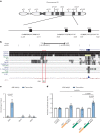
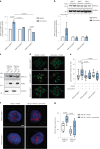
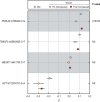
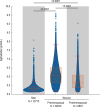
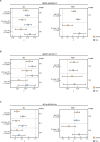
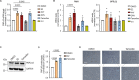
Fatty liver disease (FLD) caused by metabolic dysfunction is the leading cause of liver disease and the prevalence is rising, especially in women. Although during reproductive age women are protected against FLD, for still unknown and understudied reasons some develop rapidly progressive disease at the menopause. The patatin-like phospholipase domain-containing 3 (PNPLA3) p.I148M variant accounts for the largest fraction of inherited FLD variability. In the present study, we show that there is a specific multiplicative interaction between female sex and PNPLA3 p.I148M in determining FLD in at-risk individuals (steatosis and fibrosis, P < 10-10; advanced fibrosis/hepatocellular carcinoma, P = 0.034) and in the general population (P < 10-7 for alanine transaminase levels). In individuals with obesity, hepatic PNPLA3 expression was higher in women than in men (P = 0.007) and in mice correlated with estrogen levels. In human hepatocytes and liver organoids, PNPLA3 was induced by estrogen receptor-α (ER-α) agonists. By chromatin immunoprecipitation and luciferase assays, we identified and characterized an ER-α-binding site within a PNPLA3 enhancer and demonstrated via CRISPR-Cas9 genome editing that this sequence drives PNPLA3 p.I148M upregulation, leading to lipid droplet accumulation and fibrogenesis in three-dimensional multilineage spheroids with stellate cells. These data suggest that a functional interaction between ER-α and PNPLA3 p.I148M variant contributes to FLD in women.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no conflicts of interest relevant to the present study. L.V. has received speaking fees from MSD, Gilead, AlfaSigma and AbbVie, served as a consultant for Gilead, Pfizer, AstraZeneca, Novo Nordisk, Intercept, Diatech Pharmacogenetics, Ionis Pharmaceuticals, Boeringher Ingelheim and Resalis, and received research grants from Gilead. D.P. served as a consultant for, and has received speaking fees, travel grants and research grants from, Macopharma, Ortho Clinical Diagnostics, Grifols, Terumo, Immucor, Diamed and Diatech Pharmacogenetics.
Figures



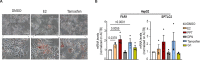

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

