Functional diversity of c-di-GMP receptors in prokaryotic and eukaryotic systems
- PMID: 37749602
- PMCID: PMC10519070
- DOI: 10.1186/s12964-023-01263-5
Functional diversity of c-di-GMP receptors in prokaryotic and eukaryotic systems
Abstract
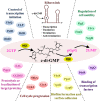
Cyclic bis-(3', 5')-dimeric guanosine monophosphate (c-di-GMP) is ubiquitous in many bacterial species, where it functions as a nucleotide-based secondary messenger and is a vital regulator of numerous biological processes. Due to its ubiquity, most bacterial species possess a wide range of downstream receptors that has a binding affinity to c-di-GMP and elicit output responses. In eukaryotes, several enzymes and riboswitches operate as receptors that interact with c-di-GMP and transduce cellular or environmental signals. This review examines the functional variety of receptors in prokaryotic and eukaryotic systems that exhibit distinct biological responses after interacting with c-di-GMP. Evolutionary relationships and similarities in distance among the c-di-GMP receptors in various bacterial species were evaluated to understand their specificities. Furthermore, residues of receptors involved in c-di-GMP binding are summarized. This review facilitates the understanding of how distinct receptors from different origins bind c-di-GMP equally well, yet fulfill diverse biological roles at the interspecies, intraspecies, and interkingdom levels. Furthermore, it also highlights c-di-GMP receptors as potential therapeutic targets, particularly those found in pathogenic microorganisms. Video Abstract.
Keywords: Bacteria; Binding affinity; Eukaryotes; Evolutionary relatedness; Interspecies; Receptor; c-di-GMP.
© 2023. BioMed Central Ltd., part of Springer Nature.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

