Capsid-host interactions for HIV-1 ingress
- PMID: 37750702
- PMCID: PMC10732038
- DOI: 10.1128/mmbr.00048-22
Capsid-host interactions for HIV-1 ingress
Abstract
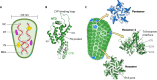
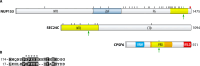
The HIV-1 capsid, composed of approximately 1,200 copies of the capsid protein, encases genomic RNA alongside viral nucleocapsid, reverse transcriptase, and integrase proteins. After cell entry, the capsid interacts with a myriad of host factors to traverse the cell cytoplasm, pass through the nuclear pore complex (NPC), and then traffic to chromosomal sites for viral DNA integration. Integration may very well require the dissolution of the capsid, but where and when this uncoating event occurs remains hotly debated. Based on size constraints, a long-prevailing view was that uncoating preceded nuclear transport, but recent research has indicated that the capsid may remain largely intact during nuclear import, with perhaps some structural remodeling required for NPC traversal. Completion of reverse transcription in the nucleus may further aid capsid uncoating. One canonical type of host factor, typified by CPSF6, leverages a Phe-Gly (FG) motif to bind capsid. Recent research has shown these peptides reside amid prion-like domains (PrLDs), which are stretches of protein sequence devoid of charged residues. Intermolecular PrLD interactions along the exterior of the capsid shell impart avid host factor binding for productive HIV-1 infection. Herein we overview capsid-host interactions implicated in HIV-1 ingress and discuss important research questions moving forward. Highlighting clinical relevance, the long-acting ultrapotent inhibitor lenacapavir, which engages the same capsid binding pocket as FG host factors, was recently approved to treat people living with HIV.
Keywords: CPSF6; FG domain; HIV; NUP153; capsid; lenacapavir; liquid-liquid phase separation; mixed-charge domain; nuclear import; prion-like domain; speckle-associated domain; trafficking.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

