Exploring avibactam and relebactam inhibition of Klebsiella pneumoniae carbapenemase D179N variant: role of the Ω loop-held deacylation water
- PMID: 37750722
- PMCID: PMC10583681
- DOI: 10.1128/aac.00350-23
Exploring avibactam and relebactam inhibition of Klebsiella pneumoniae carbapenemase D179N variant: role of the Ω loop-held deacylation water
Abstract
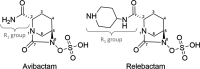
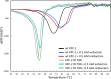
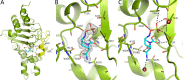
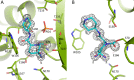
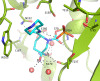
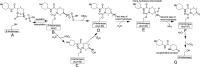
Klebsiella pneumoniae carbapenemase-2 (KPC-2) presents a clinical threat as this β-lactamase confers resistance to carbapenems. Recent variants of KPC-2 in clinical isolates contribute to concerning resistance phenotypes. Klebsiella pneumoniae expressing KPC-2 D179Y acquired resistance to the ceftazidime/avibactam combination affecting both the β-lactam and the β-lactamase inhibitor yet has lowered minimum inhibitory concentrations for all other β-lactams tested. Furthermore, Klebsiella pneumoniae expressing the KPC-2 D179N variant also manifested resistance to ceftazidime/avibactam yet retained its ability to confer resistance to carbapenems although significantly reduced. This structural study focuses on the inhibition of KPC-2 D179N by avibactam and relebactam and expands our previous analysis that examined ceftazidime resistance conferred by D179N and D179Y variants. Crystal structures of KPC-2 D179N soaked with avibactam and co-crystallized with relebactam were determined. The complex with avibactam reveals avibactam making several hydrogen bonds, including with the deacylation water held in place by Ω loop. These results could explain why the KPC-2 D179Y variant, which has a disordered Ω loop, has a decreased affinity for avibactam. The relebactam KPC-2 D179N complex revealed a new orientation of the diazabicyclooctane (DBO) intermediate with the scaffold piperidine ring rotated ~150° from the standard DBO orientation. The density shows relebactam to be desulfated and present as an imine-hydrolysis intermediate not previously observed. The tetrahedral imine moiety of relebactam interacts with the deacylation water. The rotated relebactam orientation and deacylation water interaction could potentially contribute to KPC-mediated DBO fragmentation. These results elucidate important differences that could aid in the design of novel β-lactamase inhibitors.
Keywords: antibiotic resistance; protein crystallography; β-lactamase; β-lactamase inhibitor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
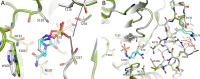
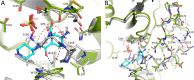
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

