Type I interferon signature and cycling lymphocytes in macrophage activation syndrome
- PMID: 37751296
- PMCID: PMC10645381
- DOI: 10.1172/JCI165616
Type I interferon signature and cycling lymphocytes in macrophage activation syndrome
Abstract
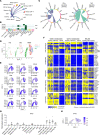
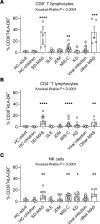
BACKGROUNDMacrophage activation syndrome (MAS) is a life-threatening complication of Still's disease (SD) characterized by overt immune cell activation and cytokine storm. We aimed to further understand the immunologic landscape of SD and MAS.METHODWe profiled PBMCs from people in a healthy control group and patients with SD with or without MAS using bulk RNA-Seq and single-cell RNA-Seq (scRNA-Seq). We validated and expanded the findings by mass cytometry, flow cytometry, and in vitro studies.RESULTSBulk RNA-Seq of PBMCs from patients with SD-associated MAS revealed strong expression of genes associated with type I interferon (IFN-I) signaling and cell proliferation, in addition to the expected IFN-γ signal, compared with people in the healthy control group and patients with SD without MAS. scRNA-Seq analysis of more than 65,000 total PBMCs confirmed IFN-I and IFN-γ signatures and localized the cell proliferation signature to cycling CD38+HLA-DR+ cells within CD4+ T cell, CD8+ T cell, and NK cell populations. CD38+HLA-DR+ lymphocytes exhibited prominent IFN-γ production, glycolysis, and mTOR signaling. Cell-cell interaction modeling suggested a network linking CD38+HLA-DR+ lymphocytes with monocytes through IFN-γ signaling. Notably, the expansion of CD38+HLA-DR+ lymphocytes in MAS was greater than in other systemic inflammatory conditions in children. In vitro stimulation of PBMCs demonstrated that IFN-I and IL-15 - both elevated in MAS patients - synergistically augmented the generation of CD38+HLA-DR+ lymphocytes, while Janus kinase inhibition mitigated this response.CONCLUSIONMAS associated with SD is characterized by overproduction of IFN-I, which may act in synergy with IL-15 to generate CD38+HLA-DR+ cycling lymphocytes that produce IFN-γ.
Keywords: Cytokines; Immunology; Inflammation.
Figures




References
-
- Petty RE, et al. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31(2):390–392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

