The catalytic subunit of DNA-PK regulates transcription and splicing of AR in advanced prostate cancer
- PMID: 37751307
- PMCID: PMC10645393
- DOI: 10.1172/JCI169200
The catalytic subunit of DNA-PK regulates transcription and splicing of AR in advanced prostate cancer
Abstract
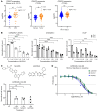
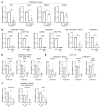
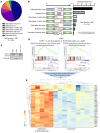
Aberrant androgen receptor (AR) signaling drives prostate cancer (PC), and it is a key therapeutic target. Although initially effective, the generation of alternatively spliced AR variants (AR-Vs) compromises efficacy of treatments. In contrast to full-length AR (AR-FL), AR-Vs constitutively activate androgenic signaling and are refractory to the current repertoire of AR-targeting therapies, which together drive disease progression. There is an unmet clinical need, therefore, to develop more durable PC therapies that can attenuate AR-V function. Exploiting the requirement of coregulatory proteins for AR-V function has the capacity to furnish tractable routes for attenuating persistent oncogenic AR signaling in advanced PC. DNA-PKcs regulates AR-FL transcriptional activity and is upregulated in both early and advanced PC. We hypothesized that DNA-PKcs is critical for AR-V function. Using a proximity biotinylation approach, we demonstrated that the DNA-PK holoenzyme is part of the AR-V7 interactome and is a key regulator of AR-V-mediated transcription and cell growth in models of advanced PC. Crucially, we provide evidence that DNA-PKcs controls global splicing and, via RBMX, regulates the maturation of AR-V and AR-FL transcripts. Ultimately, our data indicate that targeting DNA-PKcs attenuates AR-V signaling and provide evidence that DNA-PKcs blockade is an effective therapeutic option in advanced AR-V-positive patients with PC.
Keywords: Endocrinology; Oncology; Prostate cancer.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

