Structure of the bc1- cbb3 respiratory supercomplex from Pseudomonas aeruginosa
- PMID: 37751552
- PMCID: PMC10556555
- DOI: 10.1073/pnas.2307093120
Structure of the bc1- cbb3 respiratory supercomplex from Pseudomonas aeruginosa
Abstract
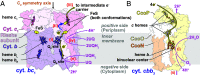
Energy conversion by electron transport chains occurs through the sequential transfer of electrons between protein complexes and intermediate electron carriers, creating the proton motive force that enables ATP synthesis and membrane transport. These protein complexes can also form higher order assemblies known as respiratory supercomplexes (SCs). The electron transport chain of the opportunistic pathogen Pseudomonas aeruginosa is closely linked with its ability to invade host tissue, tolerate harsh conditions, and resist antibiotics but is poorly characterized. Here, we determine the structure of a P. aeruginosa SC that forms between the quinol:cytochrome c oxidoreductase (cytochrome bc1) and one of the organism's terminal oxidases, cytochrome cbb3, which is found only in some bacteria. Remarkably, the SC structure also includes two intermediate electron carriers: a diheme cytochrome c4 and a single heme cytochrome c5. Together, these proteins allow electron transfer from ubiquinol in cytochrome bc1 to oxygen in cytochrome cbb3. We also present evidence that different isoforms of cytochrome cbb3 can participate in formation of this SC without changing the overall SC architecture. Incorporating these different subunit isoforms into the SC would allow the bacterium to adapt to different environmental conditions. Bioinformatic analysis focusing on structural motifs in the SC suggests that cytochrome bc1-cbb3 SCs also exist in other bacterial pathogens.
Keywords: Pseudomonas aeruginosa; cryoEM; electron transport chain; respiratory supercomplexes; structure.
Conflict of interest statement
The authors declare no competing interest.
Figures



Similar articles
-
Mobile cytochrome c2 and membrane-anchored cytochrome cy are both efficient electron donors to the cbb3- and aa3-type cytochrome c oxidases during respiratory growth of Rhodobacter sphaeroides.J Bacteriol. 2001 Mar;183(6):2013-24. doi: 10.1128/JB.183.6.2013-2024.2001. J Bacteriol. 2001. PMID: 11222600 Free PMC article.
-
Structure and redox properties of the diheme electron carrier cytochrome c4 from Pseudomonas aeruginosa.J Inorg Biochem. 2020 Feb;203:110889. doi: 10.1016/j.jinorgbio.2019.110889. Epub 2019 Oct 22. J Inorg Biochem. 2020. PMID: 31707335 Free PMC article.
-
Cryo-EM structure and kinetics reveal electron transfer by 2D diffusion of cytochrome c in the yeast III-IV respiratory supercomplex.Proc Natl Acad Sci U S A. 2021 Mar 16;118(11):e2021157118. doi: 10.1073/pnas.2021157118. Proc Natl Acad Sci U S A. 2021. PMID: 33836592 Free PMC article.
-
Terminal Respiratory Oxidases: A Targetables Vulnerability of Mycobacterial Bioenergetics?Front Cell Infect Microbiol. 2020 Nov 23;10:589318. doi: 10.3389/fcimb.2020.589318. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33330134 Free PMC article. Review.
-
Cytochrome bc1-aa3 Oxidase Supercomplex As Emerging and Potential Drug Target Against Tuberculosis.Curr Mol Pharmacol. 2022;15(2):380-392. doi: 10.2174/1874467214666210928152512. Curr Mol Pharmacol. 2022. PMID: 34602044 Review.
Cited by
-
The structure of the diheme cytochrome c4 from Neisseria gonorrhoeae reveals multiple contributors to tuning reduction potentials.J Inorg Biochem. 2024 Apr;253:112496. doi: 10.1016/j.jinorgbio.2024.112496. Epub 2024 Jan 24. J Inorg Biochem. 2024. PMID: 38330683 Free PMC article.
-
Membrane-Bound Redox Enzyme Cytochrome bd-I Promotes Carbon Monoxide-Resistant Escherichia coli Growth and Respiration.Int J Mol Sci. 2024 Jan 20;25(2):1277. doi: 10.3390/ijms25021277. Int J Mol Sci. 2024. PMID: 38279276 Free PMC article.
-
Interplay of niche and respiratory network in shaping bacterial colonization.J Biol Chem. 2025 Jan;301(1):108052. doi: 10.1016/j.jbc.2024.108052. Epub 2024 Dec 9. J Biol Chem. 2025. PMID: 39662826 Free PMC article. Review.
-
Structure of a dimeric full-length ABC transporter.Nat Commun. 2024 Nov 16;15(1):9946. doi: 10.1038/s41467-024-54147-8. Nat Commun. 2024. PMID: 39550367 Free PMC article.
-
Carbon Monoxide and Prokaryotic Energy Metabolism.Int J Mol Sci. 2025 Mar 20;26(6):2809. doi: 10.3390/ijms26062809. Int J Mol Sci. 2025. PMID: 40141451 Free PMC article. Review.
References
-
- Nicholls D. G., Ferguson S. J., Bioenergetics (Version 3.5.1., ed. 4th, Academic Press, 2013). Accessed 25 April 2023.
-
- Stuchebrukhov A., Schäfer J., Berg J., Brzezinski P., Kinetic advantage of forming respiratory supercomplexes. Biochim. Biophys. Acta 1861, 148193 (2020). - PubMed
-
- Gu J., et al. , The architecture of the mammalian respirasome. Nature 537, 639–643 (2016). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

