Generation and molecular characterization of human pluripotent stem cell-derived pharyngeal foregut endoderm
- PMID: 37751684
- PMCID: PMC10637111
- DOI: 10.1016/j.devcel.2023.08.024
Generation and molecular characterization of human pluripotent stem cell-derived pharyngeal foregut endoderm
Abstract
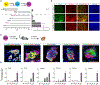
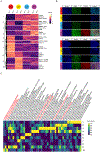
Approaches to study human pharyngeal foregut endoderm-a developmental intermediate that is linked to various human syndromes involving pharynx development and organogenesis of tissues such as thymus, parathyroid, and thyroid-have been hampered by scarcity of tissue access and cellular models. We present an efficient stepwise differentiation method to generate human pharyngeal foregut endoderm from pluripotent stem cells. We determine dose and temporal requirements of signaling pathway engagement for optimized differentiation and characterize the differentiation products on cellular and integrated molecular level. We present a computational classification tool, "CellMatch," and transcriptomic classification of differentiation products on an integrated mouse scRNA-seq developmental roadmap confirms cellular maturation. Integrated transcriptomic and chromatin analyses infer differentiation stage-specific gene regulatory networks. Our work provides the method and integrated multiomic resource for the investigation of disease-relevant loci and gene regulatory networks and their role in developmental defects affecting the pharyngeal endoderm and its derivatives.
Keywords: gene regulatory networks; human pluripotent stem cells; multiomic analyses; pharyngeal endoderm; retinoic acid pathway; stem cell differentiation.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




References
-
- Magaletta ME, Siller R, and Maehr R (2020). Differentiation of human pluripotent stem cells toward pharyngeal endoderm derivatives: Current status and potential. Current topics in developmental biology 138, 175–208. - PubMed
-
- Kearns NA, Genga RMJ, Ziller M, Peters H, Brehm MA, Meissner A, and Maehr R (2013). Generation of organized anterior foregut epithelia from pluripotent stem cells using small molecules. Stem cell research 11, 1003–1012. - PubMed
-
- Sun X, Xu J, Lu H, Liu W, Miao Z, Sui X, Liu H, Su L, Du W, He Q, et al. (2013). Directed Differentiation of Human Embryonic Stem Cells into Thymic Epithelial Progenitor-like Cells Reconstitutes the Thymic Microenvironment In Vivo. Cell stem cell 13, 230–236. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

