Machine learning for cross-scale microscopy of viruses
- PMID: 37751685
- PMCID: PMC10545915
- DOI: 10.1016/j.crmeth.2023.100557
Machine learning for cross-scale microscopy of viruses
Abstract
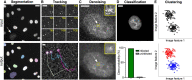
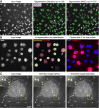
Despite advances in virological sciences and antiviral research, viruses continue to emerge, circulate, and threaten public health. We still lack a comprehensive understanding of how cells and individuals remain susceptible to infectious agents. This deficiency is in part due to the complexity of viruses, including the cell states controlling virus-host interactions. Microscopy samples distinct cellular infection stages in a multi-parametric, time-resolved manner at molecular resolution and is increasingly enhanced by machine learning and deep learning. Here we discuss how state-of-the-art artificial intelligence (AI) augments light and electron microscopy and advances virological research of cells. We describe current procedures for image denoising, object segmentation, tracking, classification, and super-resolution and showcase examples of how AI has improved the acquisition and analyses of microscopy data. The power of AI-enhanced microscopy will continue to help unravel virus infection mechanisms, develop antiviral agents, and improve viral vectors.
Keywords: CP: Microbiology and CP: Imaging; SARS-CoV-2; adenovirus tracking and trafficking; artificial intelligence; deep learning; electron microscopy; fluorescence super-resolution microscopy; herpes simplex virus; human immunodeficiency virus; influenza virus; machine learning; nanoparticle.
Copyright © 2023 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Tangye S.G., Al-Herz W., Bousfiha A., Cunningham-Rundles C., Franco J.L., Holland S.M., Klein C., Morio T., Oksenhendler E., Picard C., et al. Human Inborn Errors of Immunity: 2022 Update on the Classification from the International Union of Immunological Societies Expert Committee. J. Clin. Immunol. 2022;42:1473–1507. doi: 10.1007/s10875-022-01289-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

