Environment and taxonomy shape the genomic signature of prokaryotic extremophiles
- PMID: 37752120
- PMCID: PMC10522608
- DOI: 10.1038/s41598-023-42518-y
Environment and taxonomy shape the genomic signature of prokaryotic extremophiles
Abstract
This study provides comprehensive quantitative evidence suggesting that adaptations to extreme temperatures and pH imprint a discernible environmental component in the genomic signature of microbial extremophiles. Both supervised and unsupervised machine learning algorithms were used to analyze genomic signatures, each computed as the k-mer frequency vector of a 500 kbp DNA fragment arbitrarily selected to represent a genome. Computational experiments classified/clustered genomic signatures extracted from a curated dataset of [Formula: see text] extremophile (temperature, pH) bacteria and archaea genomes, at multiple scales of analysis, [Formula: see text]. The supervised learning resulted in high accuracies for taxonomic classifications at [Formula: see text], and medium to medium-high accuracies for environment category classifications of the same datasets at [Formula: see text]. For [Formula: see text], our findings were largely consistent with amino acid compositional biases and codon usage patterns in coding regions, previously attributed to extreme environment adaptations. The unsupervised learning of unlabelled sequences identified several exemplars of hyperthermophilic organisms with large similarities in their genomic signatures, in spite of belonging to different domains in the Tree of Life.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures


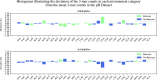
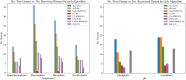
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

