PARP14 inhibition restores PD-1 immune checkpoint inhibitor response following IFNγ-driven acquired resistance in preclinical cancer models
- PMID: 37752135
- PMCID: PMC10522711
- DOI: 10.1038/s41467-023-41737-1
PARP14 inhibition restores PD-1 immune checkpoint inhibitor response following IFNγ-driven acquired resistance in preclinical cancer models
Abstract
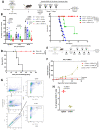
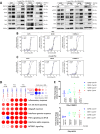
Resistance mechanisms to immune checkpoint blockade therapy (ICBT) limit its response duration and magnitude. Paradoxically, Interferon γ (IFNγ), a key cytokine for cellular immunity, can promote ICBT resistance. Using syngeneic mouse tumour models, we confirm that chronic IFNγ exposure confers resistance to immunotherapy targeting PD-1 (α-PD-1) in immunocompetent female mice. We observe upregulation of poly-ADP ribosyl polymerase 14 (PARP14) in chronic IFNγ-treated cancer cell models, in patient melanoma with elevated IFNG expression, and in melanoma cell cultures from ICBT-progressing lesions characterised by elevated IFNγ signalling. Effector T cell infiltration is enhanced in tumours derived from cells pre-treated with IFNγ in immunocompetent female mice when PARP14 is pharmacologically inhibited or knocked down, while the presence of regulatory T cells is decreased, leading to restoration of α-PD-1 sensitivity. Finally, we determine that tumours which spontaneously relapse in immunocompetent female mice following α-PD-1 therapy upregulate IFNγ signalling and can also be re-sensitised upon receiving PARP14 inhibitor treatment, establishing PARP14 as an actionable target to reverse IFNγ-driven ICBT resistance.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare the following competing interests: D.T.I., C.L., N.R.P. and M.N are all employees and shareholders of Ribon Therapeutics at the time of data collection. P.E.R. served as a consultant to Ribon Therapeutics. A.H. received research sponsorship from Ribon Therapeutics. All other authors declare no competing interests.
Figures








References
-
- Shankaran V, et al. IFNγ and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410:1107–1111. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

