Benchmarking data-driven filtering for denoising of TCRpMHC single-cell data
- PMID: 37752190
- PMCID: PMC10522655
- DOI: 10.1038/s41598-023-43048-3
Benchmarking data-driven filtering for denoising of TCRpMHC single-cell data
Abstract
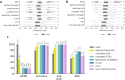
Pairing of the T cell receptor (TCR) with its cognate peptide-MHC (pMHC) is a cornerstone in T cell-mediated immunity. Recently, single-cell sequencing coupled with DNA-barcoded MHC multimer staining has enabled high-throughput studies of T cell specificities. However, the immense variability of TCR-pMHC interactions combined with the relatively low signal-to-noise ratio in the data generated using current technologies are complicating these studies. Several approaches have been proposed for denoising single-cell TCR-pMHC specificity data. Here, we present a benchmark evaluating two such denoising methods, ICON and ITRAP. We applied and evaluated the methods on publicly available immune profiling data provided by 10x Genomics. We find that both methods identified approximately 75% of the raw data as noise. We analyzed both internal metrics developed for the purpose and performance on independent data using machine learning methods trained on the raw and denoised 10x data. We find an increased signal-to-noise ratio comparing the denoised to the raw data for both methods, and demonstrate an overall superior performance of the ITRAP method in terms of both data consistency and performance. In conclusion, this study demonstrates that Improving the data quality from high throughput studies of TCRpMHC-specificity by denoising is paramount in increasing our understanding of T cell-mediated immunity.
© 2023. Springer Nature Limited.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

