The development of a highly sensitive and quantitative SARS-CoV-2 rapid antigen test applying newly developed monoclonal antibodies to an automated chemiluminescent flow-through membrane immunoassay device
- PMID: 37752417
- PMCID: PMC10523765
- DOI: 10.1186/s12865-023-00567-y
The development of a highly sensitive and quantitative SARS-CoV-2 rapid antigen test applying newly developed monoclonal antibodies to an automated chemiluminescent flow-through membrane immunoassay device
Abstract
Background: Rapid and accurate diagnosis of individuals with SARS-CoV-2 infection is an effective way to prevent and control the spread of COVID-19. Although the detection of SARS-CoV-2 viral RNA by RT-qPCR is the gold standard for COVID-19 testing, the use of antigen-detecting rapid diagnostic tests (Ag-RDTs) is emerging as a complementary surveillance tool as Omicron case numbers skyrocket worldwide. However, the results from Ag-RDTs are less accurate in individuals with low viral loads.
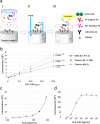
Results: To develop a highly sensitive and accurate Ag-RDT, 90 monoclonal antibodies were raised from guinea pigs immunized with SARS CoV-2 nucleocapsid protein (CoV-2-NP). By applying a capture antibody recognizing the structural epitope of the N-terminal domain of CoV-2-NP and a detection antibody recognizing the C-terminal tail of CoV-2-NP to an automated chemiluminescence flow-through membrane immunoassay device, we developed a novel Ag-RDT, CoV-2-POCube. The CoV-2-POCube exclusively recognizes CoV-2-NP variants but not the nucleocapsid proteins of other human coronaviruses. The CoV-2-POCube achieved a limit of detection sensitivity of 0.20 ~ 0.66 pg/mL of CoV-2-NPs, demonstrating more than 100 times greater sensitivity than commercially available SARS-CoV-2 Ag-RDTs.
Conclusions: CoV-2-POCube has high analytical sensitivity and can detect SARS-CoV-2 variants in 15 min without observing the high-dose hook effect, thus meeting the need for early SARS-CoV-2 diagnosis with lower viral load. CoV-2-POCube is a promising alternative to currently available diagnostic devices for faster clinical decision making in individuals with suspected COVID-19 in resource-limited settings.
Keywords: COVID-19; Lateral flow immunochromatography; Monoclonal antibody; Omicron; Point-of-care test; Rapid antigen test; SARS-CoV-2.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Comparative evaluation of RT-PCR and antigen-based rapid diagnostic tests (Ag-RDTs) for SARS-CoV-2 detection: performance, variant specificity, and clinical implications.Microbiol Spectr. 2024 Jun 4;12(6):e0007324. doi: 10.1128/spectrum.00073-24. Epub 2024 Apr 29. Microbiol Spectr. 2024. PMID: 38683014 Free PMC article.
-
Performance of rapid antigen tests to detect SARS-CoV-2 variant diversity and correlation with viral culture positivity: implication for diagnostic development and future public health strategies.mBio. 2024 Dec 11;15(12):e0273724. doi: 10.1128/mbio.02737-24. Epub 2024 Oct 31. mBio. 2024. PMID: 39480114 Free PMC article.
-
Virus variant-specific clinical performance of a SARS-CoV-2 rapid antigen test with focus on Omicron variants of concern.Clin Microbiol Infect. 2023 Aug;29(8):1085.e1-1085.e8. doi: 10.1016/j.cmi.2023.05.009. Epub 2023 May 12. Clin Microbiol Infect. 2023. PMID: 37182639 Free PMC article.
-
Accuracy of rapid point-of-care antigen-based diagnostics for SARS-CoV-2: An updated systematic review and meta-analysis with meta-regression analyzing influencing factors.PLoS Med. 2022 May 26;19(5):e1004011. doi: 10.1371/journal.pmed.1004011. eCollection 2022 May. PLoS Med. 2022. PMID: 35617375 Free PMC article.
-
Rapid antigen tests for SARS-CoV-2-a synopsis of the medical evidence.Diagn Microbiol Infect Dis. 2023 Oct;107(2):116027. doi: 10.1016/j.diagmicrobio.2023.116027. Epub 2023 Jul 14. Diagn Microbiol Infect Dis. 2023. PMID: 37516068 Review.
Cited by
-
Guinea Pigs Are Not a Suitable Model to Study Neurological Impacts of Ancestral SARS-CoV-2 Intranasal Infection.Viruses. 2025 May 15;17(5):706. doi: 10.3390/v17050706. Viruses. 2025. PMID: 40431717 Free PMC article.
References
-
- Cubuk J, Alston JJ, Incicco JJ, Singh S, Stuchell-Brereton MD, Ward MD, Zimmerman MI, Vithani N, Griffith D, Wagoner JA, et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun. 2021;12(1):1936. doi: 10.1038/s41467-021-21953-3. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

